A snakemake pipeline for the analysis of RNA-seq data that makes use of hisat2 and DESeq2.
To align, count, normalize counts and compute gene differential expressions between conditions using paired-end Illumina RNA-seq data.
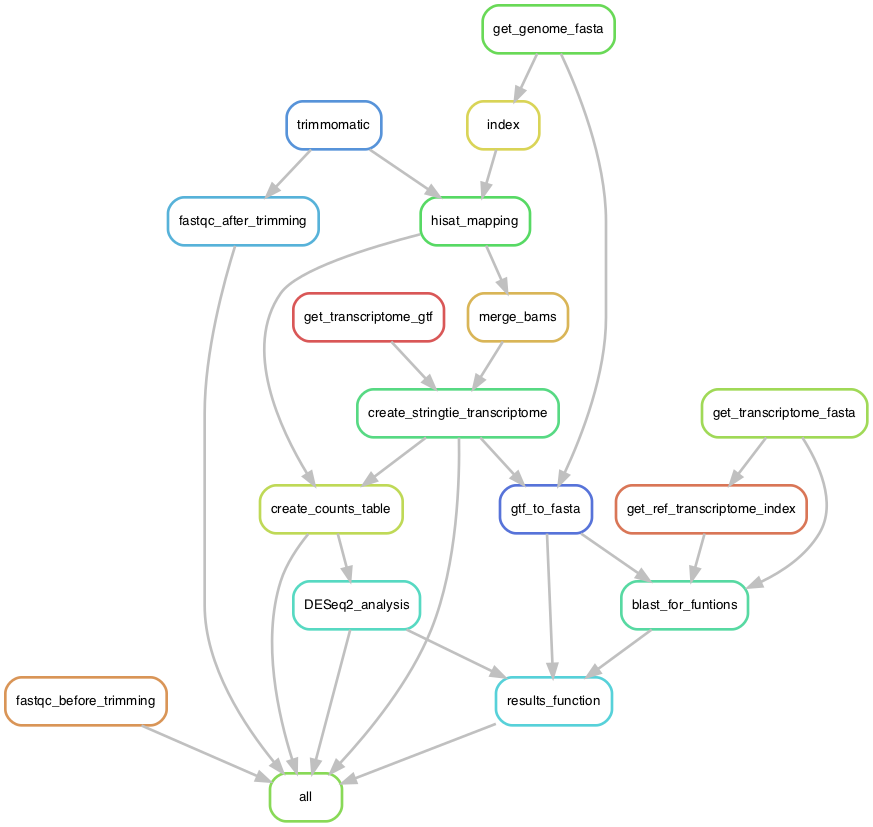
This pipeline analyses the raw RNA-seq data and produces a file containing normalized counts, differential expression, numbers of the clusters the genes have been assigned to and functions of transcripts. The raw fastq files will be trimmed for adaptors and quality checked with trimmomatic. Next, the necessary genome sequence fastas will be downloaded to be used for the mapping of the trimmed reads using hisat2. With stringtie and a downloaded reference annotation a new annotation will be created. This new annotation will be used to obtain the raw counts and do a local blast to a reference transcriptome fasta containing predicted functions. The counts are normalized and differential expressions are calculated using DESeq2. significantly expressed genes are used to create heatmaps and plots both in total and clustered. finally the DESeq2, blast and clustering data are combined to get the final results table.
- Some command of the Unix Shell to connect to a remote server where you will execute the pipeline (e.g. SURF Lisa Cluster). You can find a good tutorial from the Software Carpentry Foundation here and another one from Berlin Bioinformatics here.
- Some command of the Unix Shell to transfer datasets to and from a remote server (to transfer sequencing files and retrieve the results/). The Berlin Bioinformatics Unix begginer guide available [here] should be sufficient for that (check the
wgetandscpcommands). - An understanding of the steps of a canonical RNA-Seq analysis (trimming, alignment, etc.). You can find some info here.
Snakefile: a master file that contains the desired outputs and the rules to generate them from the input files.config.yaml: the configuration files making the Snakefile adaptable to any input files, genome and parameter for the rules.data/: a folder containing samples.txt (sample descriptions) and subsetted paired-end fastq files used to test locally the pipeline. Generated using Seqtk:seqtk sample -s100 {inputfile(can be gzipped)} 250000 > {output(always gunzipped)}This folder should contain thefastqof the paired-end RNA-seq data, you want to run.envs/: a folder containing the environments needed for the conda package manager. If run with the--use-condacommand, Snakemake will install the necessary softwares and packages using the conda environment files.samples.tsv: a file containing information about the names, the paths and the conditions of the samples used as input for the pipeline. This file has to be adapted to your sample names before running the pipeline.
You will need a local copy of the Snakemake_hisat-DESeq on your machine. You can either:
- use git in the shell:
git clone git@github.com:KoesGroup/Snakemake_hisat-DESeq.git - click on "Clone or download" and select
download
First, you need to create an environment where Snakemake and the python pandaspackage will be installed. To do that, we will use the conda package manager.
- Create a virtual environment named
rnasequsing theglobal_env.yamlfile with the following command:conda env create --name rnaseq --file envs/global_env.yamlThen, activate this virtual environment withsource activate rnaseq
The Snakefile will then take care of installing and loading the packages and softwares required by each step of the pipeline.
Make sure you have changed the parameters in the config.yaml file that specifies where to find the sample data file, the genomic and transcriptomic reference fasta files to use and the parameters for certains rules etc.
This file is used so the Snakefile does not need to be changed when locations or parameters need to be changed.
The Snakemake pipeline/workflow management system reads a master file (often called Snakefile) to list the steps to be executed and defining their order. It has many rich features. Read more here.
From the folder containing the Snakefile, use the command snakemake --use-conda -np to perform a dry run that prints out the rules and commands.
Simply type Snakemake --use-conda and provide the number of cores with --cores 10 for ten cores for instance.
For cluster execution, please refer to the Snakemake reference.
-
the RNA-Seq read alignment files *.bam
-
the fastqc report files *.html
-
the unscaled RNA-Seq read counts: counts.txt
-
the differential expression file results.tsv
-
the combined results file final.txt