scramblyzer consists of several modules (see their list using scramblyzer -h) performing various analyses connected to lipid scrambling (lipid flip-flops).
Module composition calculates lipid composition of a membrane (in time, if requested).
Module positions calculates position of each lipid head in time.
Module rate calculates the percentage of scrambled lipids in time.
Module flipflops calculates the number of flip-flop events that occured during the simulations.
scramblyzer requires you to have groan library installed. You can get groan from here. See also the installation instructions for groan.
- Run
make groan=PATH_TO_GROANto create a binary filescramblyzerthat you can place wherever you want.PATH_TO_GROANis a path to the directory containing groan library (containinggroan.handlibgroan.a). - (Optional) Run
make installto copy the the binary filescramblyzerinto${HOME}/.local/bin.
Usage: scramblyzer MODULE OPTIONS
MODULES
composition calculates lipid composition of a membrane
positions calculates position of each lipid head in time
rate calculates percentage of scrambled lipids in time
flipflops calculates the number of flip-flop events
Note that in all the modules, atoms can be selected using the groan selection language.
As scramblyzer is primarily designed for the analysis of Martini simulations, it natively recognizes all standard (and some non-standard) Martini lipids (over 200 lipid types). You can also add your own lipids by supplying a file lipids.txt into the directory from which you call scramblyzer. In this file, scramblyzer expects one lipid type (residue name) per line. The maximal length of the lipid name is 4 characters. The lipids.txt file may contain comments initiated by #. The maximal length of each line is 1023 characters.
You can also add any additional lipids directly into the scramblyzer code (by modifying the variable default_lipid_names in the function read_lipid_names located in the file src/general.c) and recompiling the program using make groan=PATH_TO_GROAN.
Valid OPTIONS for the composition module:
-h print this message and exit
-c STRING gro file to read
-f STRING xtc file to read (optional)
-n STRING ndx file to read (optional, default: index.ndx)
-o STRING output file name (default: composition.xvg)
-p STRING selection of lipid head identifiers (default: name PO4)
-t FLOAT time interval between analyzed trajectory frames in ns (default: 1.0)
Note that the options -o and -t are only used when xtc file is provided (flag -f). Otherwise the results are written to standard output (i.e. terminal).
scramblyzer composition -c md.gro
Scramblyzer will calculate lipid composition of a membrane based on the atoms and their coordinates from md.gro. Lipids will be assigned to the upper or lower leaflet based on the position of their 'lipid head identifiers' (in this case the selection name PO4, default option of the flag -p) relative to the center of all identified lipid atoms. Note that scramblyzer expects one 'lipid head identifier' atom per lipid molecule.
The output of this analysis for a POPC:DOPE:POPI membrane can look for example like this:
Lipid | Upper | Lower | Full
DOPE | 139 | 137 | 276
POPI | 69 | 68 | 137
POPC | 255 | 251 | 506
-----------------------------
TOTAL | 463 | 456 | 919
scramblyzer composition -c md.gro -f md.xtc -p "name PO4 PO1" -t 5
The program will calculate lipid composition of a membrane in time, based on the coordinates from md.xtc (the coordinates from md.gro will not be used). Lipids will be assigned to the upper or lower leaflet as described above and this assignment will be performed for every analyzed frame (in this case for trajectory frames every 5 ns, as set by the flag -t).
In this case, the 'lipid head identifiers' are defined as any atoms with the name PO4 or PO1. Note that scramblyzer still assumes that there is only one 'lipid head identifier' atom per lipid molecule.
The output of this analysis will be written into composition.xvg (default option) and can be visualized using xmgrace (xgmrace -nxy composition.xvg).
Gets the z-coordinate for each specified atom in each specified trajectory frame and writes it into an output file.
Valid OPTIONS for the positions module:
-h print this message and exit
-c STRING gro file to read
-f STRING xtc file to read
-n STRING ndx file to read (optional, default: index.ndx)
-o STRING output file name (default: positions.xvg)
-p STRING selection of lipid head identifiers (default: name PO4)
-t FLOAT time interval between analyzed frames [in ns] (default: 1.0)
scramblyzer positions -c md.gro -f md.xtc -t 5
The program will get z-coordinates of all atoms with atom name PO4 (default option of the flag -p) for frames every 5 ns (flag -t) and write these coordinates into positions.xvg (default option of the flag -o). The output file can be visualized using xmgrace (xmgrace -nxy positions.xvg).
Module rate calculates the 'scrambling rate' for individual lipid types, i.e. how often the lipids flip between the membrane leaflets.
The program will first assign lipid molecules into their respective membrane leaflets (as with scramblyzer composition) based on their coordinates in the first frame of the xtc file. This assignment will be then treated as 'reference assignment'. In all further analyzed trajectory frames, lipid molecules will be assigned to the membrane leaflets again. Then, for each lipid, the current assignment of the lipid will be compared to the 'reference assignment'. In case the lipid is currently assigned to the opposite membrane leaflet than in the 'reference assignment', it is classified as 'scrambled'. For each analyzed trajectory frame, the proportion of 'scrambled lipids' is then calculated and written into the output file.
This approach to quantifying lipid scrambling has its advantages and disadvantages. It is quite robust and insensitive to user-defined parameters (unlike the approach used in the flipflops module) but it slightly underestimates the true number of scrambling events as any lipid that flips across the membrane an even number of times will be quantified as 'not scrambled'. When analyzing very long simulations of scramblases, the calculated 'scrambling rate' will eventually converge to 50% and then fluctuate around this value not increasing further despite the fact that the lipids will still be moving between the membrane leaflets.
Valid OPTIONS for the rate module:
-h print this message and exit
-c STRING gro file to read
-f STRING xtc file to read
-n STRING ndx file to read (optional, default: index.ndx)
-o STRING output file name (default: rate.xvg)
-p STRING selection of lipid head identifiers (default: name PO4)
-t FLOAT time interval between analyzed trajectory frames in ns (default: 10.0)
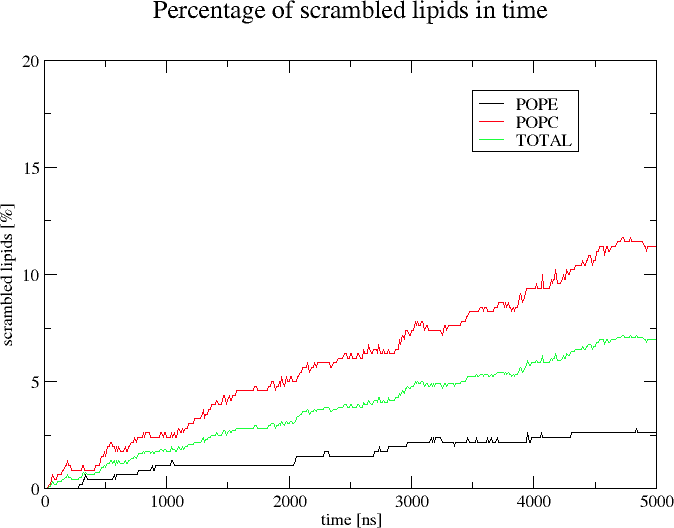
scramblyzer rate -c md.gro -f md.xtc -o scrambling.xvg
The program will calculate scrambling rate for all known lipids based on the coordinates in md.xtc (the coordinates from md.gro will not be used). A trajectory frame will be analyzed every 10 ns (default option). The 'lipid head identifiers' are the atoms named PO4 (default option) and scramblyzer expects one such atom per lipid molecule.
The scrambling rate will be calculated independently for every identified lipid type.
The output will be written into scrambling.xvg (flag -o) and can be visualized using xmgrace (xmgrace -nxy scrambling.xvg).
The plotted result for a POPC:POPE membrane containing a scramblase can look for example like this:
Module flipflops calculates the number of flip-flop events during the simulation, distinguishing flips from the upper to the lower leaflet and in the opposite direction.
The program tracks the position of every lipid head relative to the membrane center in time. If the lipid head moves across the membrane center into the other leaflet, the number of flip-flop events is increased.
The problem with this approach to quantifying lipid scrambling is the difficulty of deciding whether a particular change in the position of the lipid can be characterized as a 'flip-flop event' or whether it is merely a fluctation in the position of the lipid that is currently translocating through the membrane. In other words, if we considered any movement of the lipid head from below the membrane center to above the membrane center to be a 'flip-flop event' then (depending on our temporal resolution) a single flip-flop event could actually be counted as multiple flip-flop events. This is because during the process of lipid flip-flop, the position of the lipid head tends to fluctuate and the head may cross the membrane center multiple times. Decreasing the temporal resolution of the analysis helps with this problem but it also brings an increased risk of missing a flip-flop event that is 'real' but short-lived.
The flipflops module tries to tackle this challenge by introducing both temporal limit as well as spatial limit for the flip-flop events. For a flip-flop event to be counted by the program, the lipid head must move sufficiently far into the other membrane leaflet (spatial limit) and it must stay in this leaflet for a sufficiently long time (temporal limit). Still, deciding what values to use for these two options to obtain information about the number of 'real' flip-flop events may be difficult. The default values used by the flipflops module are by no means the only correct ones and they should not be trusted blindly and without critical evaluation. It may be better to use the module rate for a scrambling rate quantification that is less sensitive to user-defined options.
Valid OPTIONS for the flipflops module:
-h print this message and exit
-c STRING gro file to read
-f STRING xtc file to read
-n STRING ndx file to read (optional, default: index.ndx)
-p STRING selection of lipid head identifiers (default: name PO4)
-s FLOAT how far into a leaflet must the head of the lipid move to count as flip-flop [in nm] (default: 1.5)
-t INTEGER how long must the lipid stay in a leaflet to count as flip-flop [in ns] (default: 10)
scramblyzer flipflops -c md.gro -f md.xtc -s 1.75
The program will track the positions of 'lipid head identifiers' of all known lipids calculating the number of flip-flop events for these lipids based on the coordinates in md.xtc (the coordinates from md.gro will not be used). The 'lipid head identifiers' are the atoms named PO4 (default option) and scramblyzer expects one such atom per lipid molecule.
A change in the position of the 'lipid head identifier' will only be counted as a flip-flop event if the 'lipid head identifier' moves at least 1.75 nm (on the z-axis) away from the geometric center of the membrane into the opposite membrane leaflet (flag -s) and then stays in this leaflet (above or below the geometric center of the membrane, respectively) for at least 10 ns (default option). 'Membrane' is defined as all atoms of all known lipids.
The flip-flop events will be calculated separately for the individual flip-flop directions and lipid types.
The output of this analysis for a POPC:POPE membrane containing a scramblase can look for example like this:
Lipid | U->L | L->U | All
POPE | 4 | 1 | 5
POPC | 10 | 9 | 19
-----------------------------
TOTAL | 14 | 10 | 24
U->L denotes the number of flip-flop events from the upper to the lower leaflet. L->U denotes the number of flip-flop events from the lower to the upper leaflet.
Assumes that the bilayer has been built in the xy-plane (i.e. the bilayer normal is oriented along the z-axis).
scramblyzer will NOT provide reliable results when applied to simulations with curved bilayers or vesicles.
Assumes that the simulation box is rectangular and that periodic boundary conditions are applied in all three dimensions.
Note that scramblyzer always uses center of geometry, not center of mass.
Only tested on Linux. Probably will not work on anything that is not UNIX-like.