Arxiv • Installation • Training • Cite
BrLP-overview-compressed.mp4
Note
🎉 BrLP has been early-accepted and selected for oral presentation at MICCAI 2024 (top 4%)!
Download the repository, cd into the project folder and install the brlp package:
pip install -e .We recommend using a separate environment (see Anaconda). The code has been tested using python 3.9, however we expect it to work also with newer versions.
Check out our document on Data preparation and study reproducibility. This file will guide you in organizing your data and creating the required CSV files to run the training pipelines.
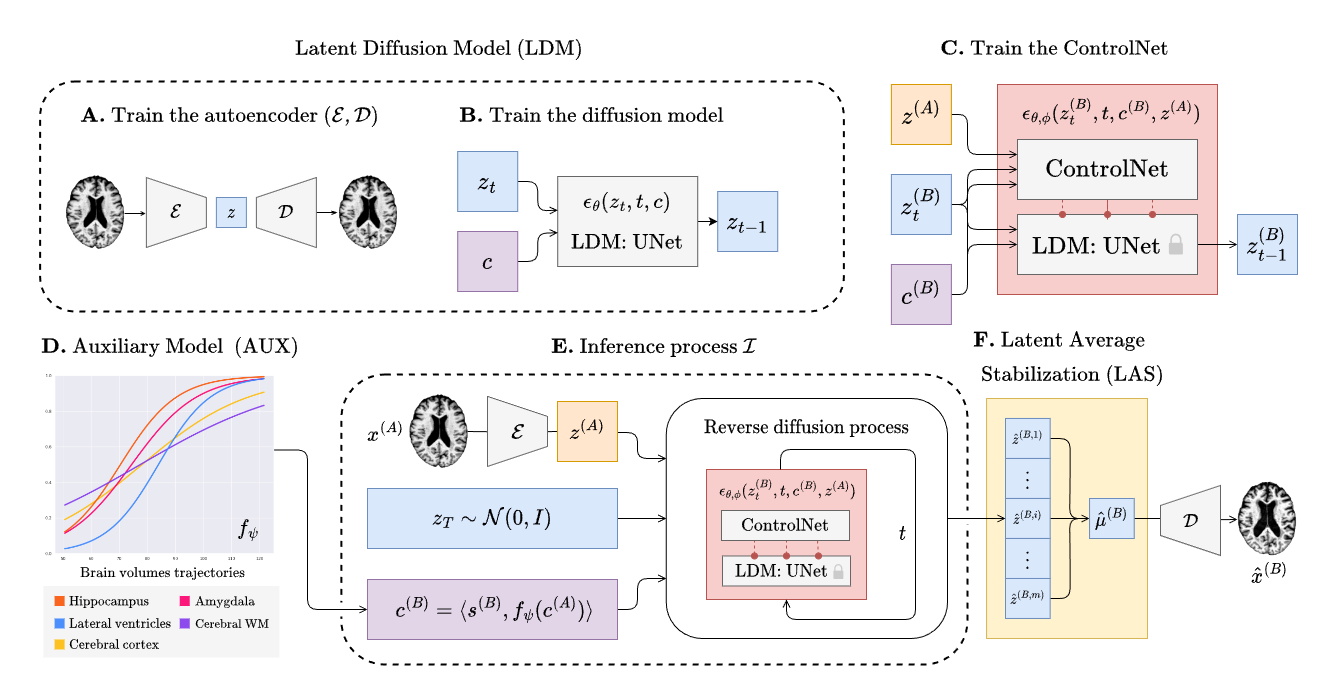
Training BrLP has 3 main phases that will be discribed in the subsequent sections. Every training (except for the auxiliary model) can be monitored using tensorboard as follows:
tensorboard --logdir runsFollow the commands below to train the autoencoder.
# Create an output and a cache directory
mkdir ae_output ae_cache
# Run the training script
python scripts/training/train_autoencoder.py \
--dataset_csv /path/to/A.csv \
--cache_dir ./ae_cache \
--output_dir ./ae_outputThen extract the latents from your MRI data:
python scripts/prepare/extract_latents.py \
--dataset_csv /path/to/A.csv \
--aekl_ckpt ae_output/autoencoder-ep-XXX.pthReplace XXX to select the autoencoder checkpoints of your choice.
Follow the commands below to train the diffusion UNet. Replace XXX to select the autoencoder checkpoints of your choice.
# Create an output and a cache directory:
mkdir unet_output unet_cache
# Run the training script
python scripts/training/train_diffusion_unet.py \
--dataset_csv /path/to/A.csv \
--cache_dir unet_cache \
--output_dir unet_output \
--aekl_ckpt ae_output/autoencoder-ep-XXX.pthFollow the commands below to train the ControlNet. Replace XXX to select the autoencoder and UNet checkpoints of your choice.
# Create an output and a cache directory:
mkdir cnet_output cnet_cache
# Run the training script
python scripts/training/train_controlnet.py \
--dataset_csv /path/to/B.csv \
--cache_dir unet_cache \
--output_dir unet_output \
--aekl_ckpt ae_output/autoencoder-ep-XXX.pth \
--diff_ckpt unet_output/unet-ep-XXX.pthFollow the commands below to train the DCM auxiliary model.
# Create an output directory
mkdir aux_output
# Run the training script
python scripts/training/train_aux.py \
--dataset_csv /path/to/A.csv \
--output_path aux_outputWe emphasize that any disease progression model capable of predicting volumetric changes over time is also viable as an auxiliary model for BrLP.
Our package comes with a brlp command to use BrLP for inference. Check:
brlp --helpThe --input parameter requires a CSV file where you list all available data for your subjects. For an example, check to examples/input.example.csv. If you haven't segmented your input scans, brlp can perform this task for you using SynthSeg, but it requires that FreeSurfer >= 7.4 be installed. The --confs parameter specifies the paths to the models and other inference parameters, such as LAS examples/confs.example.yaml.
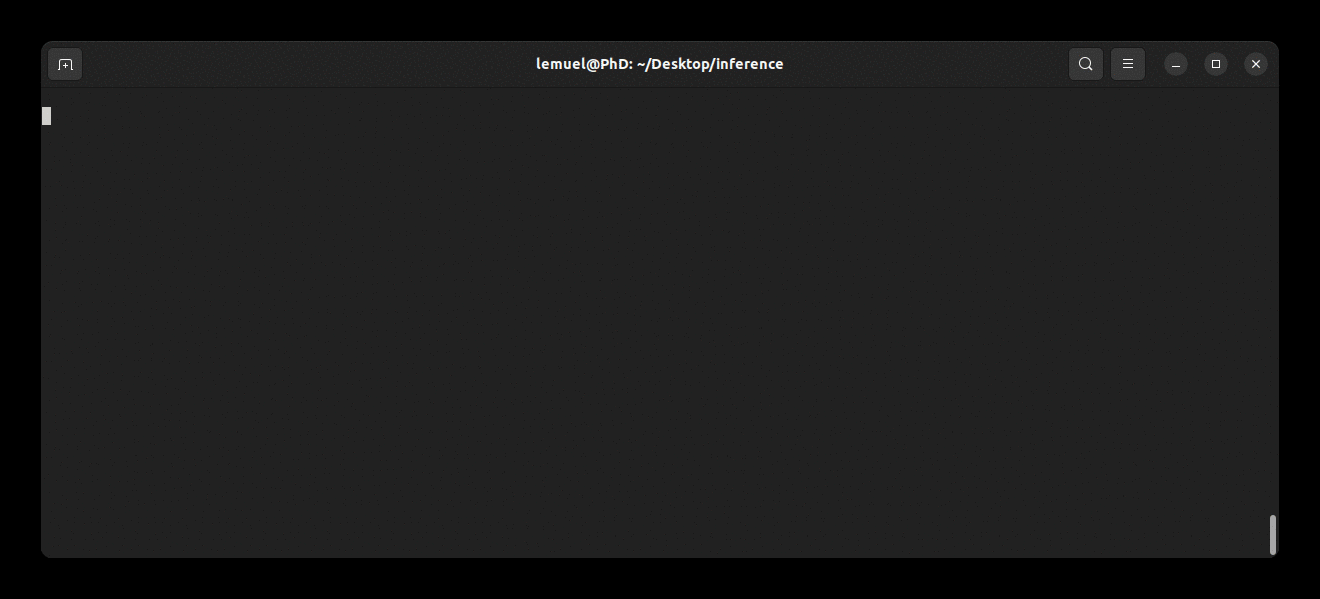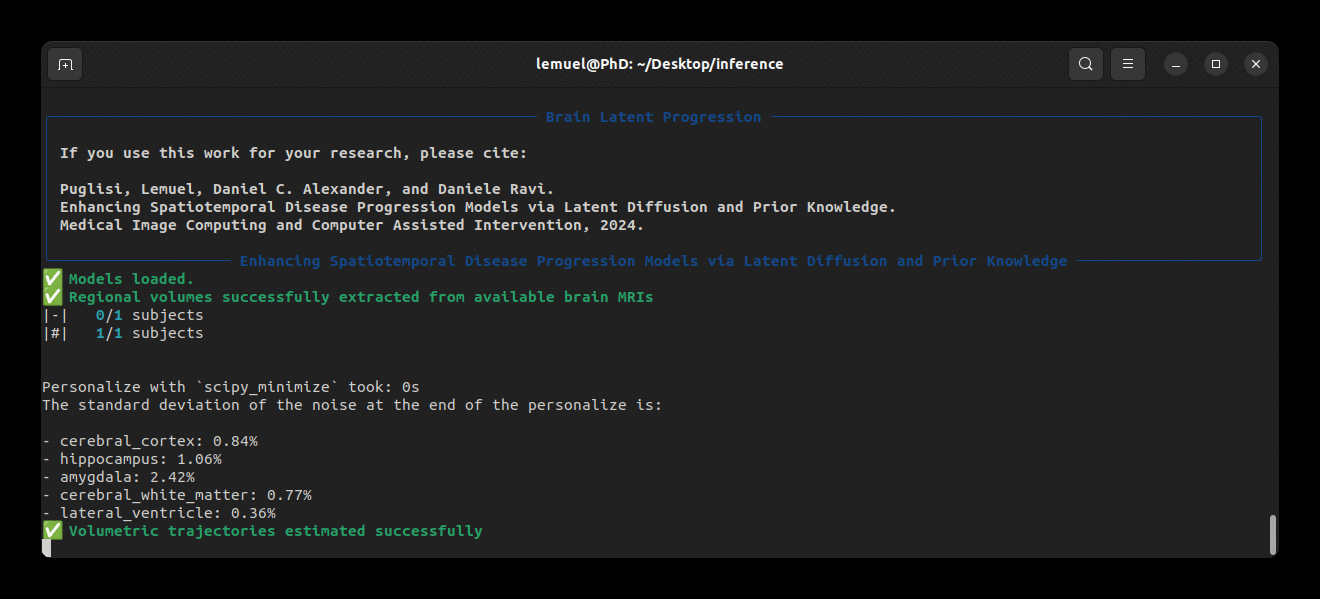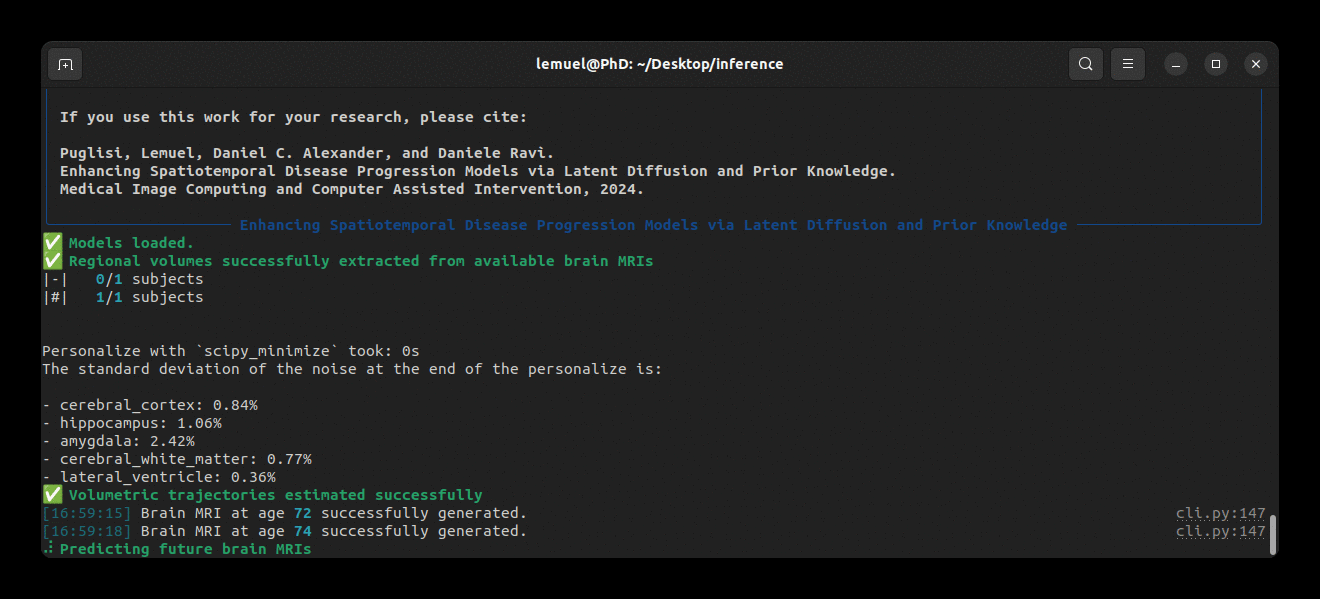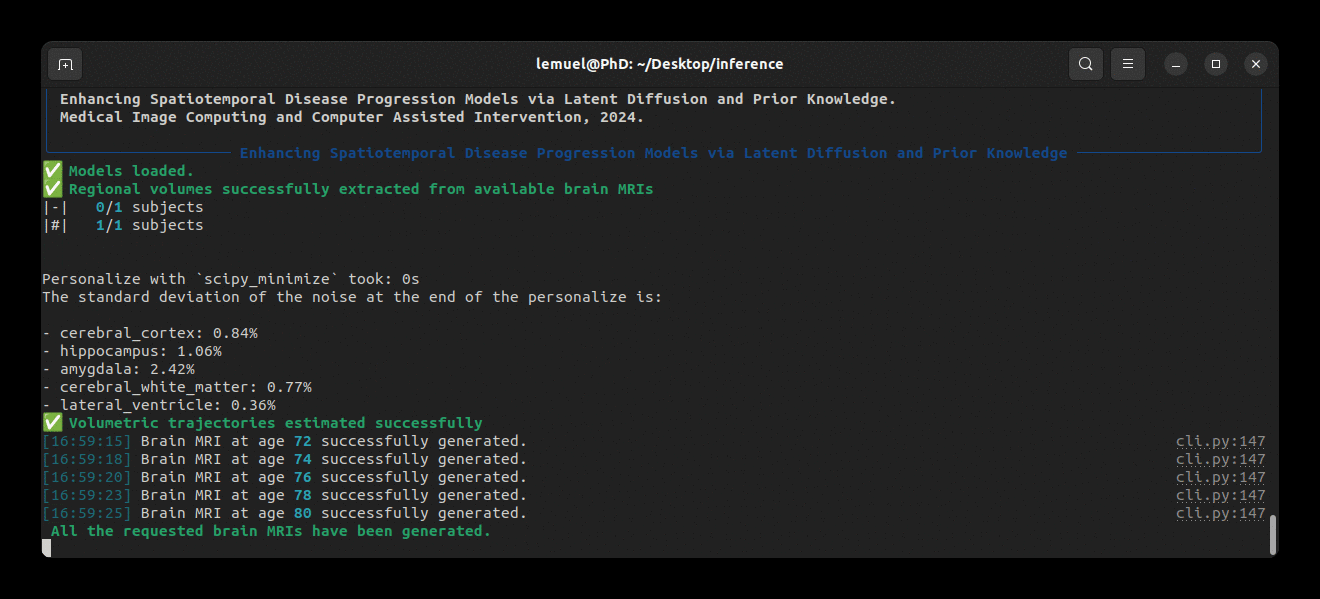
Running the program looks like this:
The pretrained models will be published upon acceptance.
| Model | Weights URL |
|---|---|
| Autoencoder | - |
| Diffusion Model UNet | - |
| ControlNet | - |
We thank the maintainers of open-source libraries for their contributions to accelerating the research process, with a special mention of MONAI and its GenerativeModels extension.
Arxiv Preprint:
@article{puglisi2024enhancing,
title={Enhancing Spatiotemporal Disease Progression Models via Latent Diffusion and Prior Knowledge},
author={Puglisi, Lemuel and Alexander, Daniel C and Rav{\`\i}, Daniele},
journal={arXiv preprint arXiv:2405.03328},
year={2024}
}MICCAI 2024 proceedings:
Coming soon.