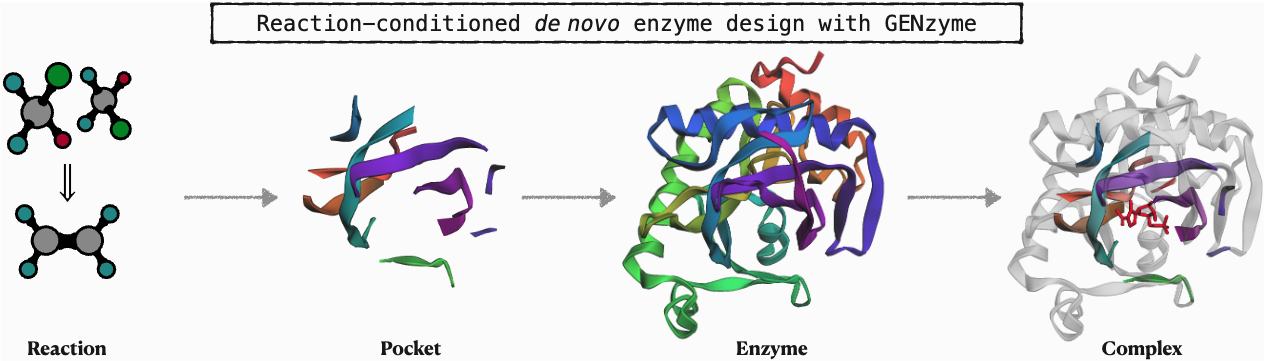
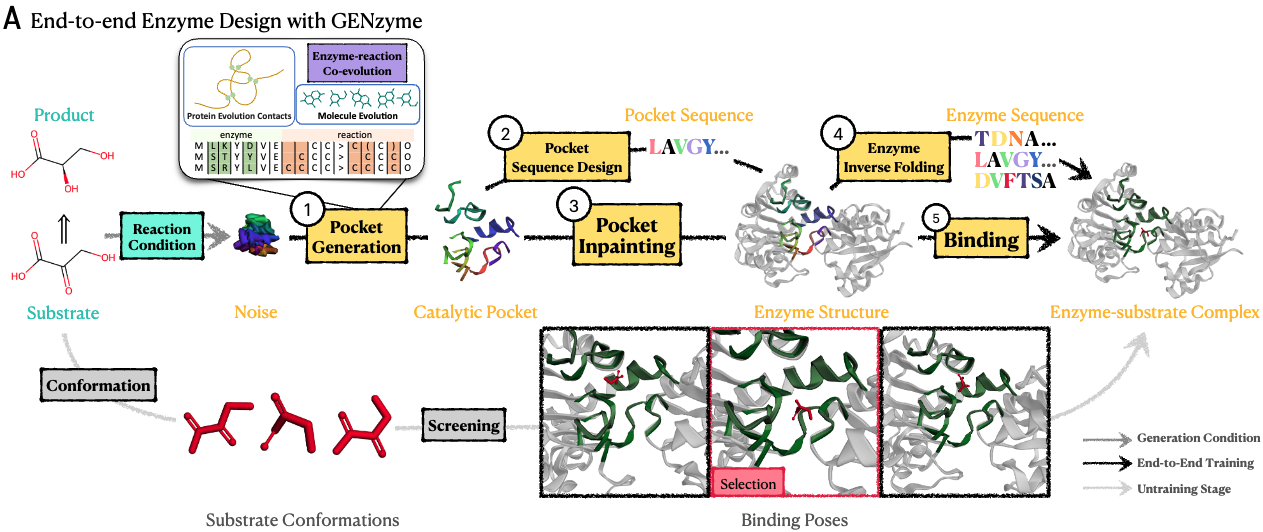
GENzyme enables de novo design of catalytic pockets, enzymes, and enzyme-substrate complexes for any reaction. Simply change args.substrate_smiles and args.product_smiles in gen_configs.py to customized substrate SMILES and product SMILES, then run python generate.py, you can design your own enzymes.
GENzyme Paper at arxiv.
python>=3.11
CUDA=12.1
torch==2.4.1 (>=2.0.0)
torch_geometric==2.4.0
torch_scatter==2.1.2
pip install mdtraj==1.10.0 (do first will install numpy, scipy as well, install later might raise dependency issues)
pip install esm==3.0.7.post1
pip install pytorch-warmup==0.1.1
pip install POT==0.9.4
pip install rdkit==2023.9.5
pip install biopython==1.84
pip install tmtools==0.2.0
pip install geomstats==2.7.0
pip install dm-tree==0.1.8
pip install ml_collections==0.1.1
pip install torchmetrics==0.11.4
pip install OpenMM
pip install einx
pip install einops
conda install conda-forge::pdbfixer
In case if you want to use the pocket-specific binding module, which is not necessarily installed for enzyme design:
For binding module, we use UniMol Docking v2, you need to install [UniCore](https://github.com/dptech-corp/Uni-Core)
You should download GENzyme checkpoint at Google drive. Once you download it, put it under genzyme_ckpt folder, namely genzyme_ckpt/genzyme.ckpt.
- Please make sure you have ESM3 installed and have access to ESM3.
- To customize catalytic reaction, remeber to change the subsrtate SMILES and product SMILES in
gen_configs.py. - You may also change
args.ptm_filterandargs.plddt_filteringen_configs.pyfor filtering enzymes. - GENzyme inference script
generate.pyis provided for your own design.
args.pdb_name #Enzyme PDB file for refinement/repurposing, set None if no PDB file available
args.substrate_smiles #Input substrate SMILES
args.product_smiles #Input product SMILES
args.n_pocket_res #Number of catalytic pocket residues for design
args.n_protein_res #Number of enzyme residues for design
args.num_pocket_design_t #Number of inference steps (ODE steps for sampling)
args.n_sample_enzyme #Number of enzymes
args.num_inpaint_t #Number of pocket inpainting steps
args.ptm_filter #pTM filtering
args.plddt_filter #pLDDT filtering
gen_configs.pycontain all inference configurations and hyperparameters.- Put your pocket pdb file under
data/ground_truth/pocket/folder, put protein pdb file underdata/ground_truth/protein/folder. - In
gen_configs.py, changeargs.pdb_nameto your pdb file name. Also changeargs.substrate_smilesto one substrate SMILES, andargs.product_smilesto one product SMILES, to customize reaction. - Run
python generate.pyfor enzyme refinement and repurposing. - Output pockets and enzymes are saved under
generated/folder.
gen_configs.pycontain all inference configurations and hyperparameters.- In
gen_configs.py, changeargs.pdb_nameto one pdb file (set to None for de novo designargs.pdb_name = None). Also changeargs.substrate_smilesto one substrate SMILES, andargs.product_smilesto one product SMILES, to customize reaction. - Run
python generate.pyfor de novo enzyme design. - Output pockets and enzymes are saved under
generated/folder.
- GENzyme reproduce script
reproduce.pyis provided. - Run
python reproduce.pyfor reproduction.
-
configs.pycontain all training configurations and hyperparameters. -
Train model using
train.pyfor single GPU training. Runpython train.pyfor training.
No Commercial use of either the model nor generated data, details to be found in LICENSE.
@misc{hua2024reactionconditionednovoenzymedesign,
title={Reaction-conditioned De Novo Enzyme Design with GENzyme},
author={Chenqing Hua and Jiarui Lu and Yong Liu and Odin Zhang and Jian Tang and Rex Ying and Wengong Jin and Guy Wolf and Doina Precup and Shuangjia Zheng},
year={2024},
eprint={2411.16694},
archivePrefix={arXiv},
primaryClass={q-bio.BM},
url={https://arxiv.org/abs/2411.16694},
}