MEG (and possibly later fMRI) analysis scripts for study group at Adv. Cognitive Neuroscience, Aarhus University.
For each group:
- Expects two numpy .npy files
- trials: with shape
(trials, sensors, time points). - labels: with shape
(trials)
- trials: with shape
- This file should be put in
data/group_x/. - Then you run
precompute_time_point_data.py(change variables first). - Now you can run either
single_time_point_analysis.py, for testing the code on a single time point, orall_time_points_analysis.py, for repeated cross-validation of each time point. This can take quite a while. - You can define custom model functions in
models.py. - Results are put in
results/time_point_models/single/group_x/. - You can use
plot_time_point_results.Rto plot your results. The images will be saved as a .tif file, which you can use directly or convert to png.
While there is code for running leave-one-group-out cross-validation, it is quite slow and doesn't seem to get good results. It's possible that some kind of windowed analysis (e.g. blocks of 10 time points) could work better.
For help: Please ask questions in an issue here on GitHub so everyone can see the answer :)
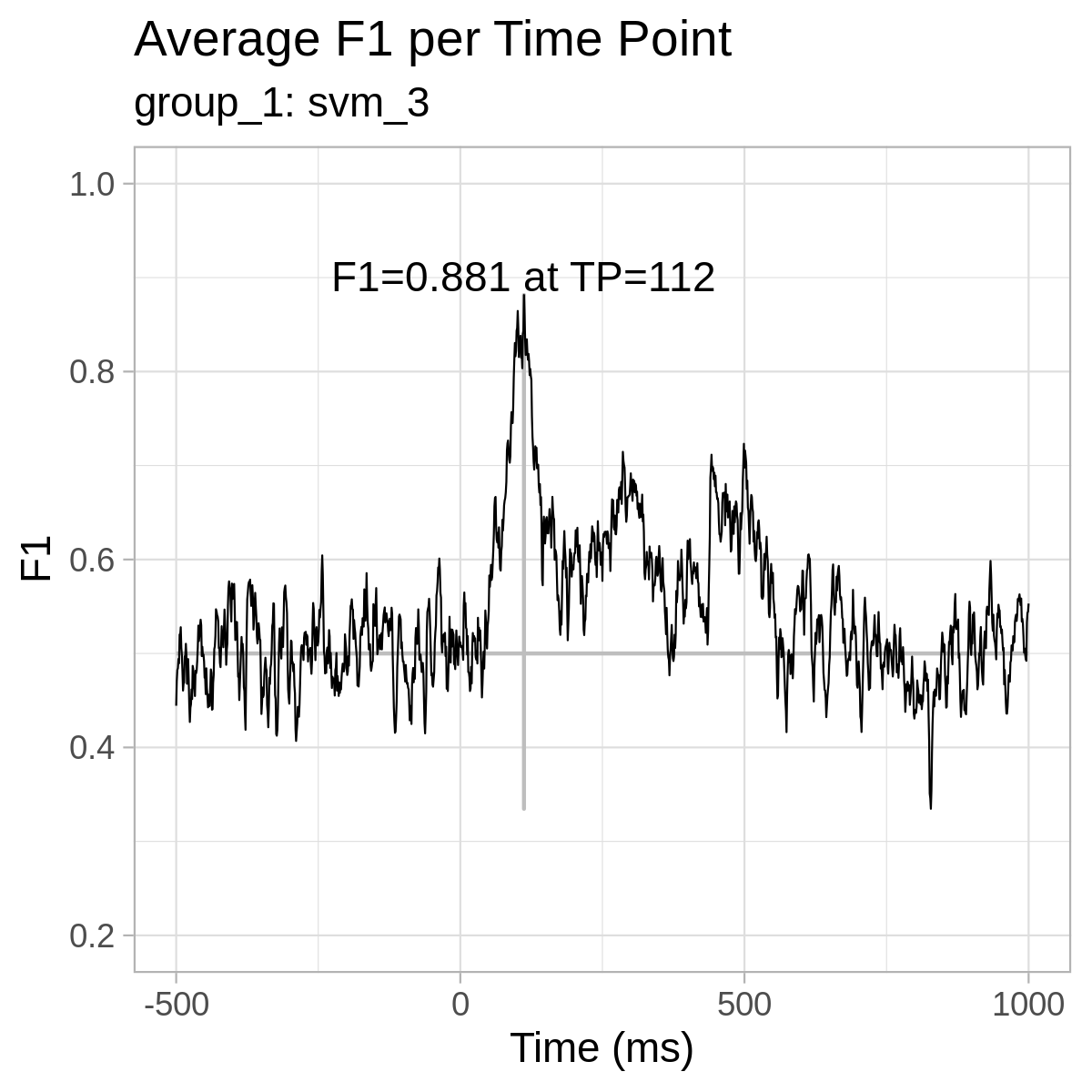
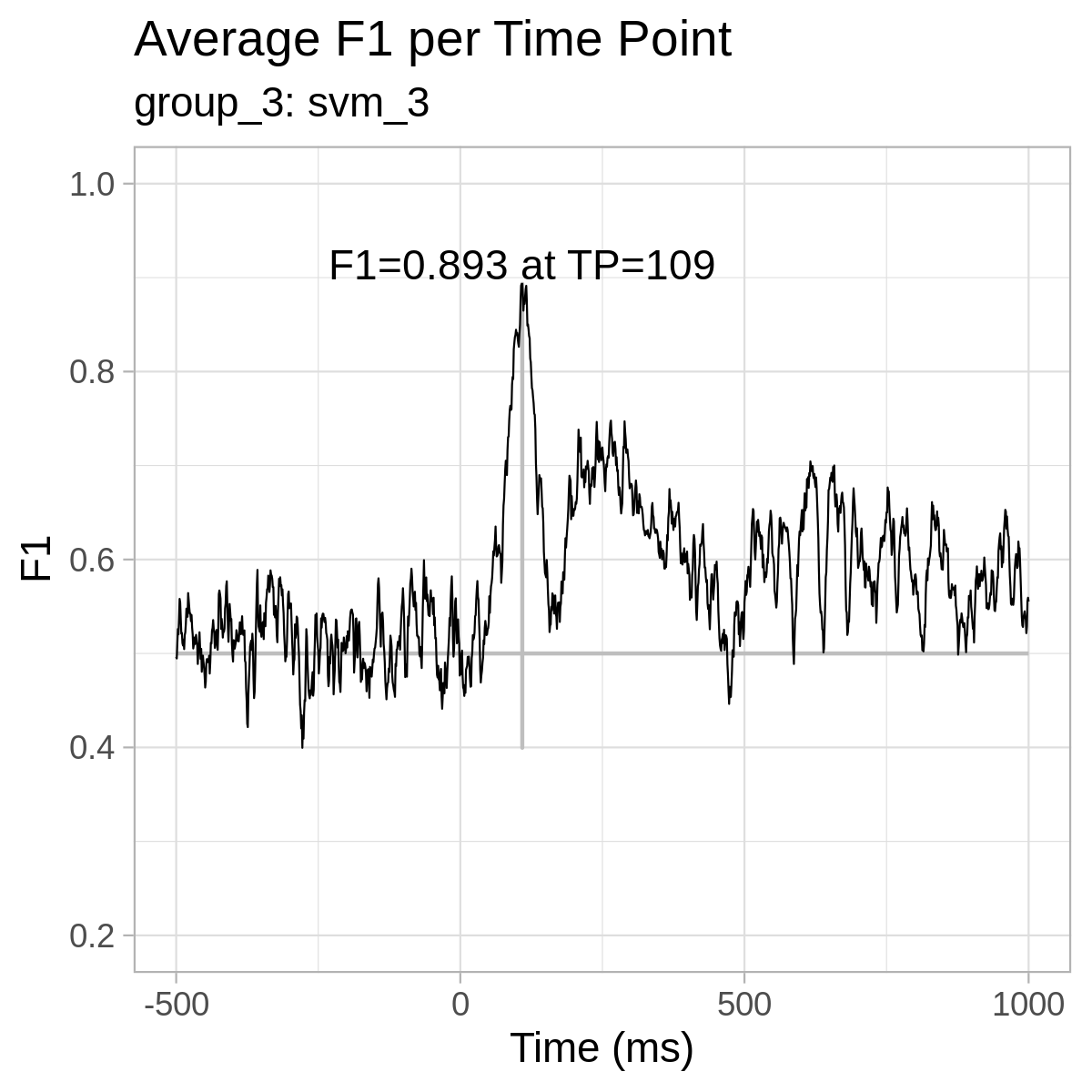
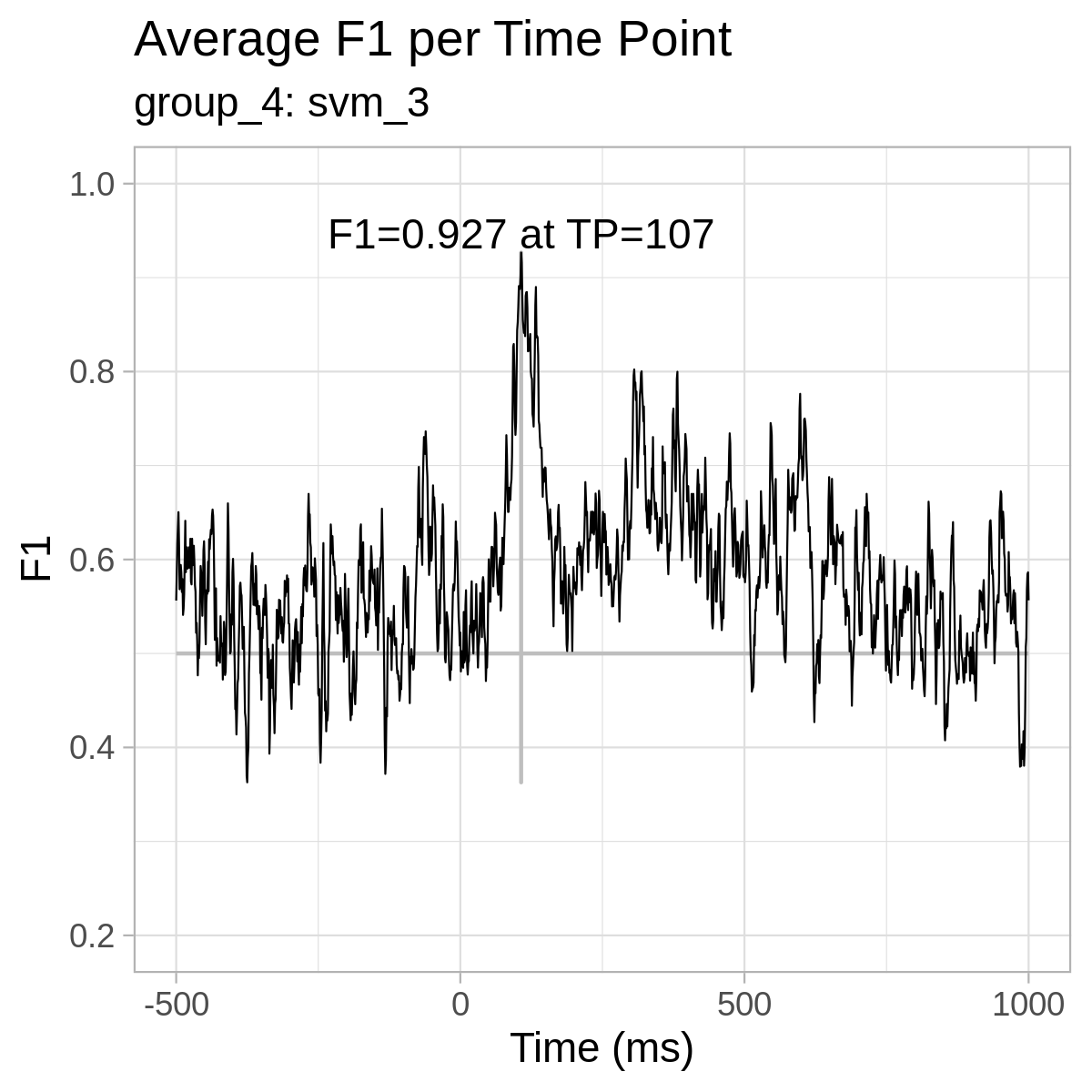
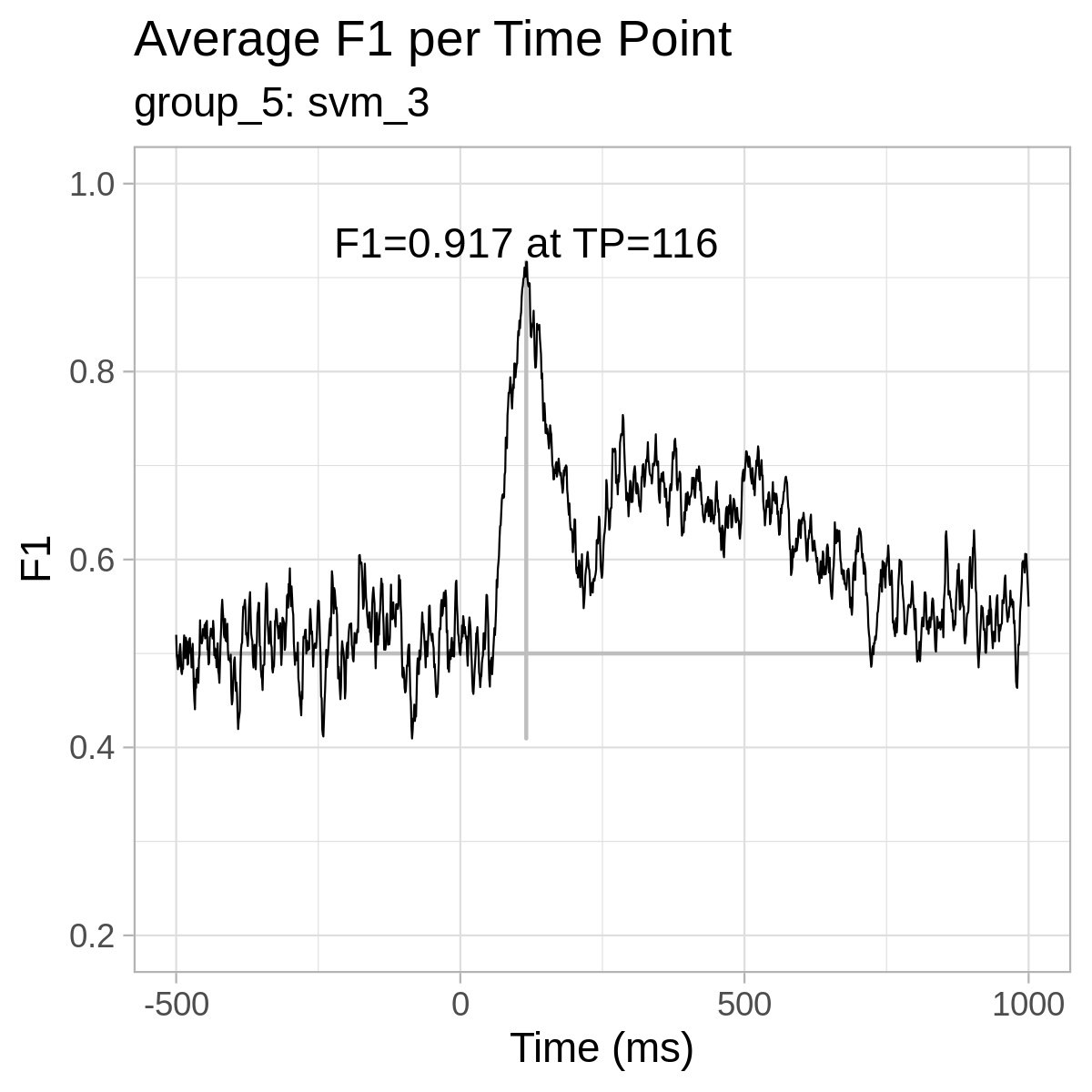
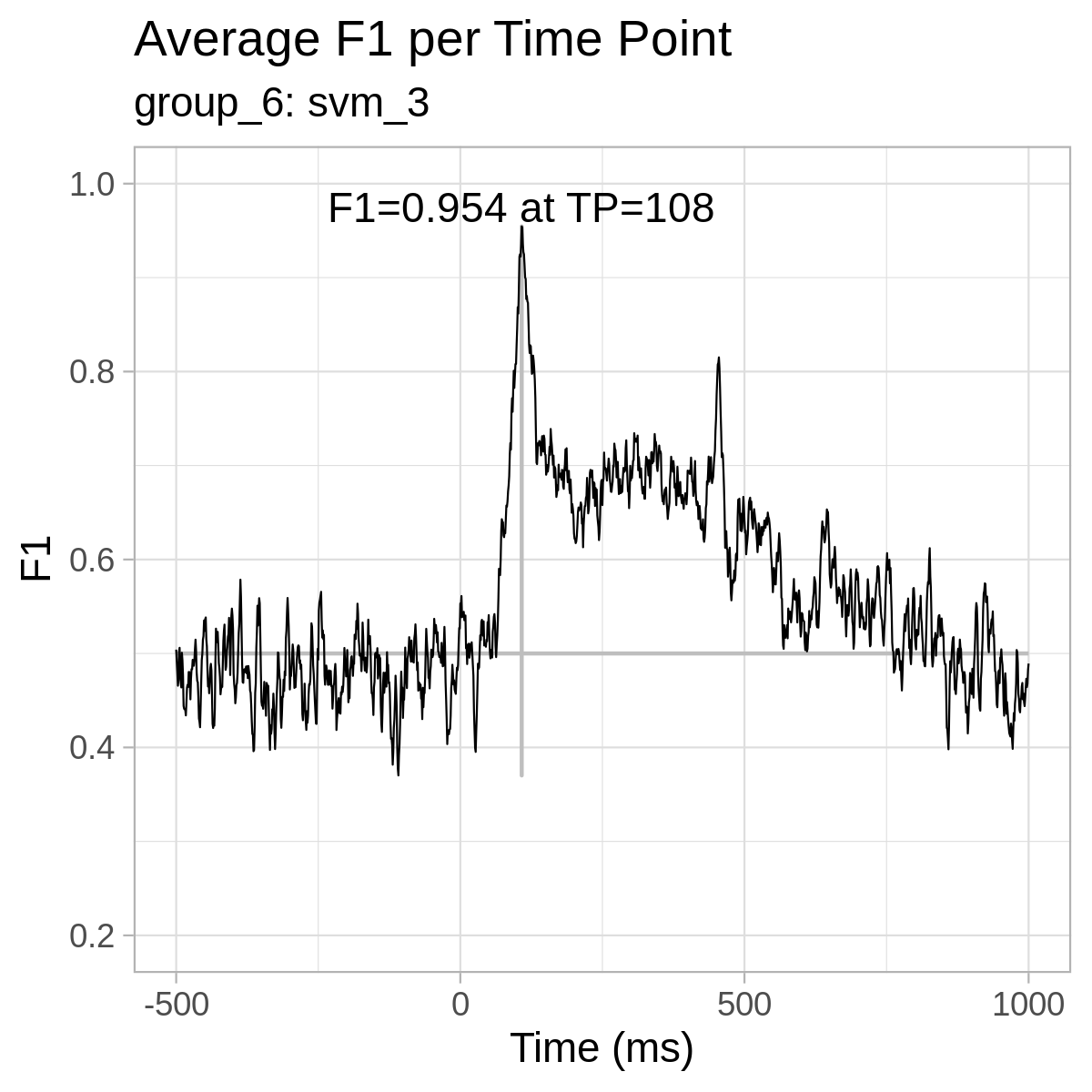
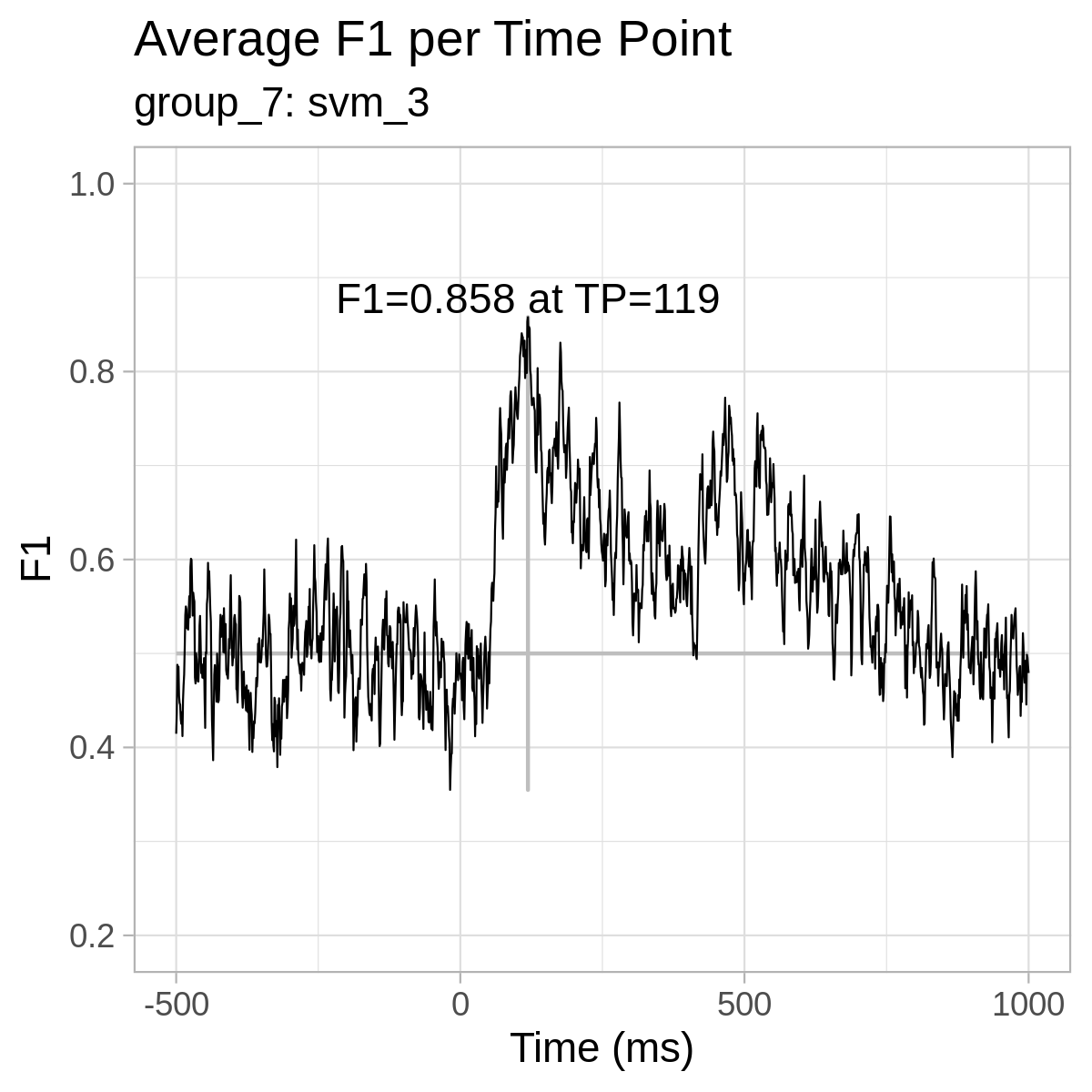
For reference only, here are the plots from our results: