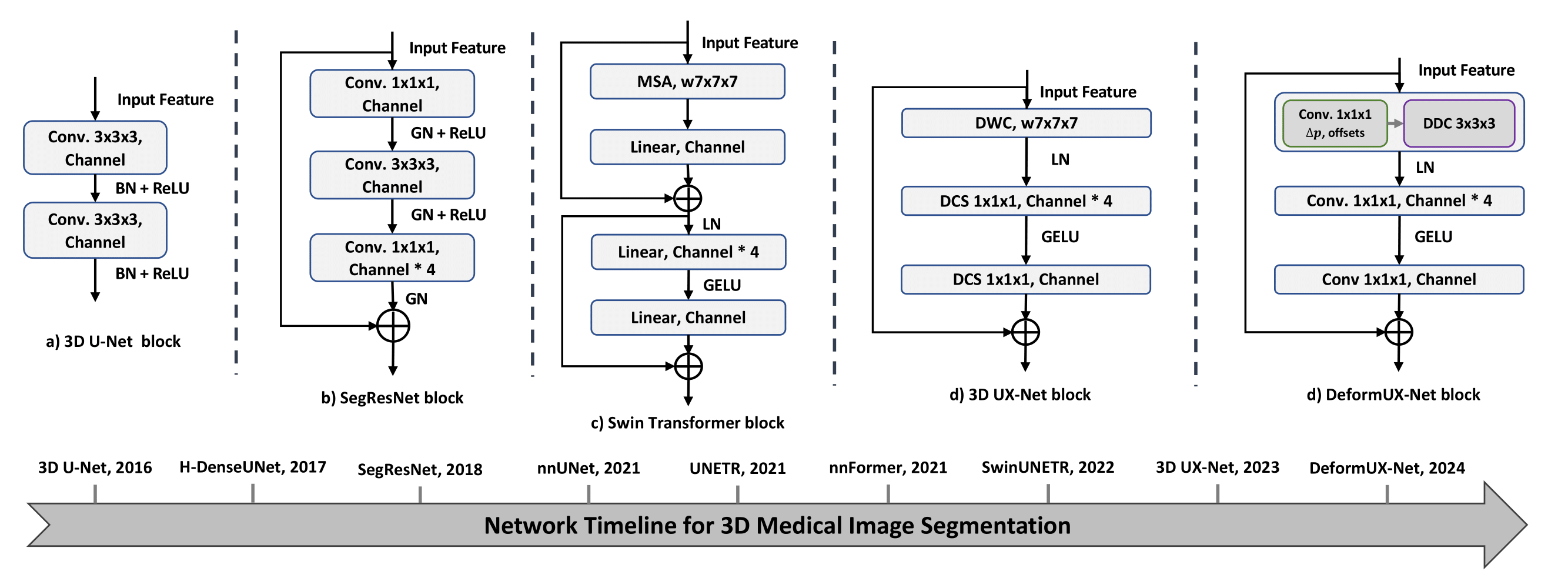
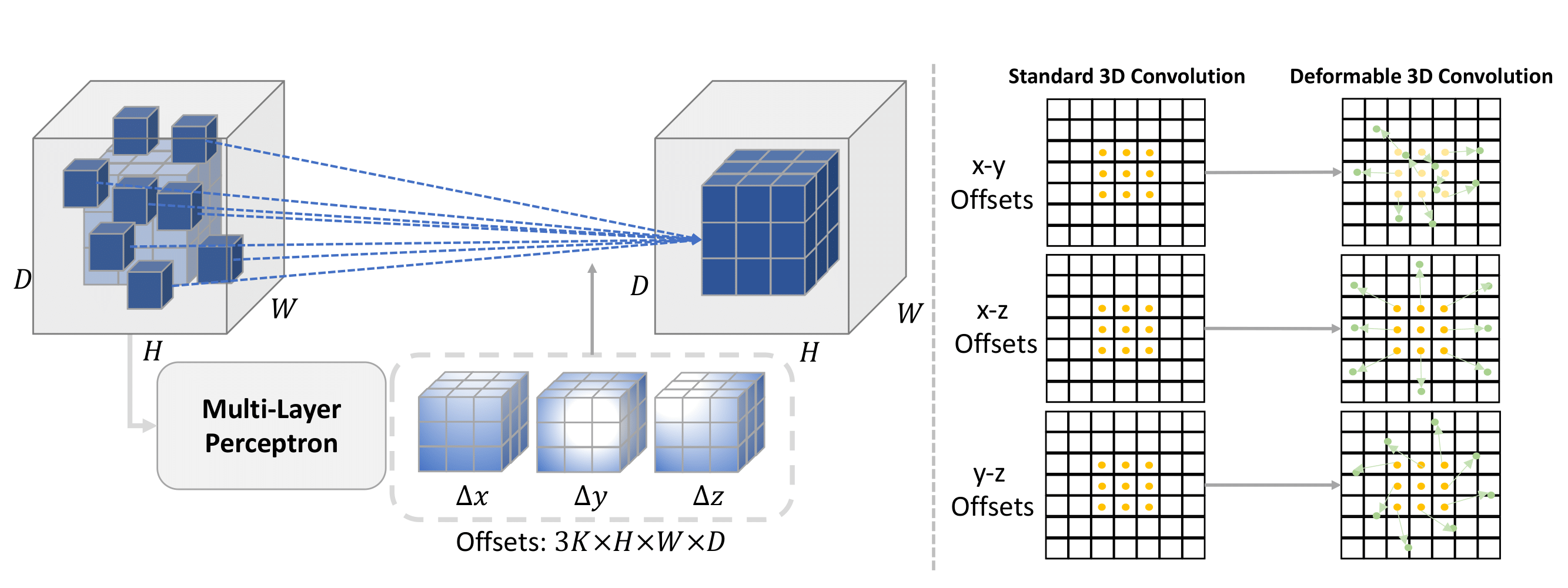
DeformUX-Net: Exploring a 3D Foundation Backbone for Medical Image Segmentation with Depthwise Deformable Convolution
Official Pytorch implementation of 3D DeformUX-Net, from the following paper:
DeformUX-Net: Exploring a 3D Foundation Backbone for Medical Image Segmentation with Depthwise Deformable Convolution. ArXiv 2023
Ho Hin Lee, Quan Liu, Qi Yang, Xin Yu, Shunxing Bao, Yuankai Huo, Bennet A. Landman
Vanderbilt University
[arXiv]
We propose 3D DeformUX-Net, a pionneering volumetric convolutional network that adapt depthwise deformable convolution as the foundation backbone to address challenges from convolution to global-local self-attention mechanisms (e.g., glbal-local range dependency trade-off, adaptive spatial aggregation, computation efficiency) for medical image segmentation.
Installation
Please look into the INSTALL.md for creating conda environment and package installation procedures.
Training Tutorial
- AMOS 2022 Training Code TRAINING.md
Results
Quantitative Performance on AMOS for Multi-Organ Segmentation (T.F.S: Train From Scratch)
| Methods | resolution | #params | FLOPs | Mean Dice (T.F.S) | Model |
|---|---|---|---|---|---|
| nn-UNet | 96x96x96 | 31.2M | 743.3G | 0.850 | |
| TransBTS | 96x96x96 | 31.6M | 110.4G | 0.783 | |
| UNETR | 96x96x96 | 92.8M | 82.6G | 0.740 | |
| nnFormer | 96x96x96 | 149.3M | 240.2G | 0.785 | |
| SwinUNETR | 96x96x96 | 62.2M | 328.4G | 0.871 | |
| 3D UX-Net (k=7) | 96x96x96 | 53.0M | 639.4G | 0.890 | |
| UNesT (Base) | 96x96x96 | 87.2M | 258.4G | 0.891 | |
| UNesT (Large) | 96x96x96 | 279.5M | 597.8G | 0.891 | |
| 3D RepUX-Net | 96x96x96 | 65.8M | 757.4G | 0.902 | |
| 3D DeformUX-Net | 96x96x96 | 55.8M | 635.8G | 0.908 |
Quantitative Performance on KiTS, MSD Pancreas, MSD Hepatic Vessels with Five-Fold Cross-Validations
| Methods | KiTS Kidney | MSD Pancreas | MSD Hepatic Vessels |
|---|---|---|---|
| nn-UNet | 0.706 | 0.703 | 0.660 |
| TransBTS | 0.669 | 0.679 | 0.613 |
| UNETR | 0.648 | 0.667 | 0.590 |
| nnFormer | 0.664 | 0.686 | 0.613 |
| SwinUNETR | 0.680 | 0.708 | 0.635 |
| 3D UX-Net (k=7) | 0.697 | 0.676 | 0.652 |
| UNesT (Base) | 0.710 | 0.690 | 0.640 |
| 3D DeformUX-Net | 0.720 | 0.717 | 0.671 |
Training
Training instructions are in TRAINING.md. Pretrained model weights will be uploaded for public usage later on.
Evaluation
Efficient evaulation can be performed for the above three public datasets as follows:
python test_seg.py --root path_to_image_folder --output path_to_output \
--dataset flare --network DEFORMUXNET --trained_weights path_to_trained_weights \
--mode test --sw_batch_size 4 --overlap 0.7 --gpu 0 --cache_rate 0.2 \
Acknowledgement
This repository is built using the timm library.
License
This project is released under the MIT license. Please see the LICENSE file for more information.
Citation
If you find this repository helpful, please consider citing:
@Article{lee2023scaling,
author = {Lee, Ho Hin and Liu, Quan and Yang, Qi and Yu, Xin and Bao, Shunxing and Huo, Yuankai and Landman, Bennett A},
title = {DeformUX-Net: Exploring a 3D Foundation Backbone for Medical Image Segmentation with Depthwise Deformable Convolution},
journal = {arXiv preprint arXiv:2310.00199},
year = {2023}
}