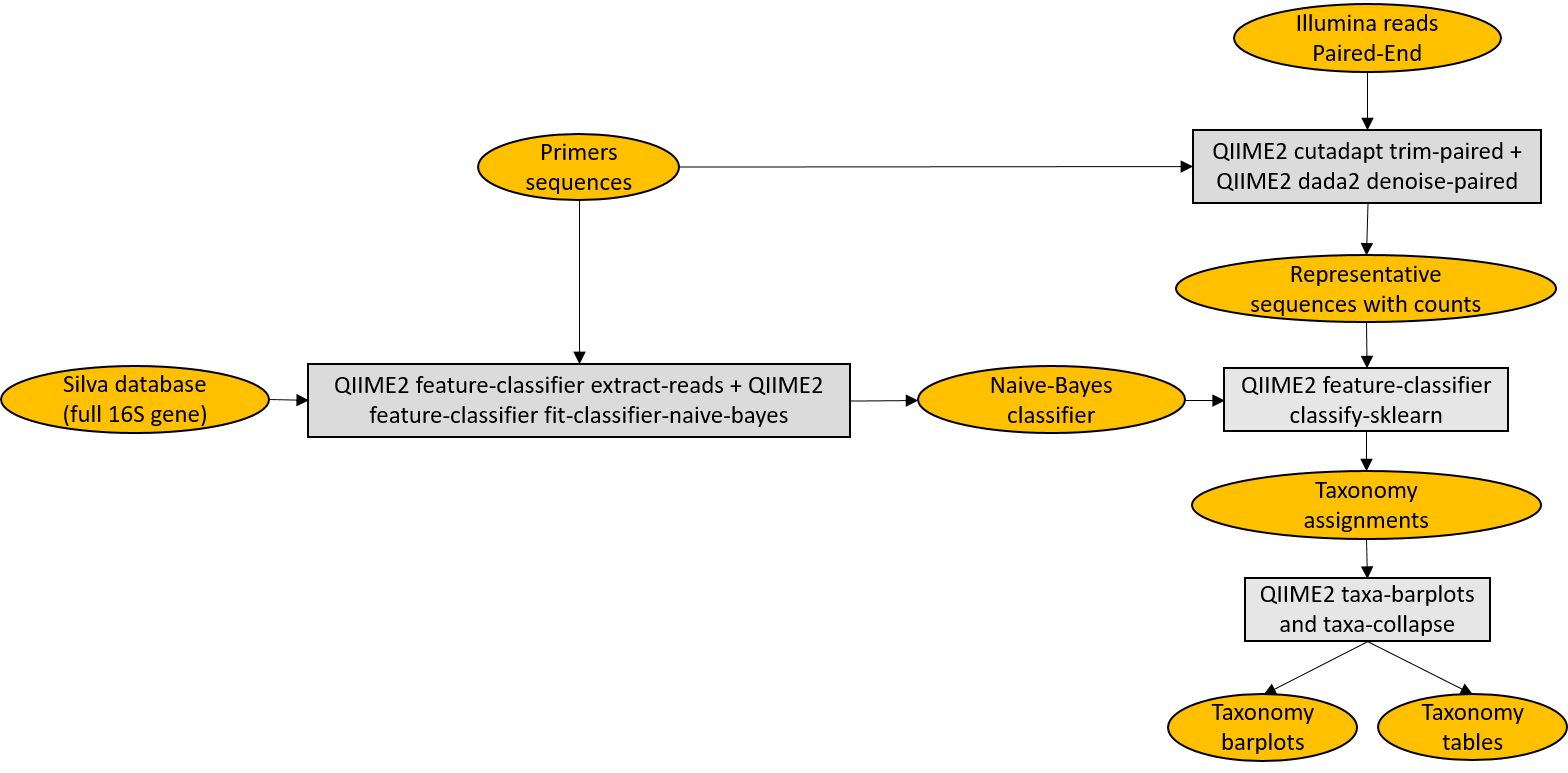
QIIME2_Illumina is a Meta-barcoding pipeline for analysing Illumina data in QIIME2 framework. Tested with Ubuntu 20.04.1 LTS.
Prerequisites
- Miniconda3.
Tested with conda 4.8.5.
which condashould return the path to the executable. If you don't have Miniconda3 installed, you could download and install it with:
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
chmod 755 Miniconda3-latest-Linux-x86_64.sh
./Miniconda3-latest-Linux-x86_64.sh
- A fasta file that you want to use as a reference database and a text file with taxonmy, or a preformatted marker gene reference database.
Installation
git clone https://github.com/MaestSi/QIIME2_Illumina.git
cd QIIME2_Illumina
chmod 755 *
./install.sh
A conda environment named QIIME2_Illumina_env is created, where qiime2-2021.8 is installed.
The QIIME2_Illumina pipeline is composed of a set of scripts which should be run sequentially. The pipeline assumes you keep the default name to output files, and leave them in the working directory.
Train_classifier.sh
Usage: Train_classifier.sh <FW_primer> <RV_primer> <DB_FASTA> <TAXONOMY_TSV> <MIN_LEN> <MAX_LEN>
Note: this script should be run only if you don't have a Naive-Bayes classifier trained on the region of interest of your marker gene yet.
Inputs:
- <FW_primer>: the sequence of the forward PCR primer
- <RV_primer>: the sequence of the reverse PCR primer
- <DB_fasta>: a fasta file containing sequences of the reference database
- <TAXONOMY_tsv>: a text file containing taxonomy corresponding to sequences in the reference database
- <MIN_LEN>: minimum expected amplicon size
- <MAX_LEN>: maximum expected amplicon size
Outputs:
- <"DB".nb-classifier.qza>: Naive-Bayes classifier trained on the region of interest of the marker gene
- <"DB".qza>: QIIME2 artifact of type DNAFASTAFormat containing reference sequences
- <"TAXONOMY".qza>: QIIME2 artifact of type HeaderlessTSVTaxonomyFormat containing taxonomy of reference sequences
Create_manifest.sh
Usage: Create_manifest.sh <sample_metadata> <reads_dir>
Inputs:
- <sample-metadata.tsv>: file containing metadata for all samples, validated with Keemei
- <reads_dir>: directory containing R1 and R2 reads in fastq.gz format
Outputs:
- <manifest.txt>: file used for importing reads in QIIME2
Import_data.sh
Usage: Import_data.sh <manifest.txt> <FW_primer> <RV_primer>
Inputs:
- <manifest.txt>: file used for importing reads in QIIME2
- <FW_primer>: the sequence of the forward PCR primer
- <RV_primer>: the sequence of the reverse PCR primer
Outputs:
- <sequences_untrimmed.qza>: reads before PCR primers trimming
- <sequences.qza>: reads after PCR primers trimming
- <demux_summary_untrimmed.qzv>: QIIME2 visualization file for inspecting sequencing quality before PCR primers trimming
- <demux_summary.qzv>: QIIME2 visualization file for inspecting sequencing quality after PCR primers trimming
Denoise_sequences.sh
Usage: Denoise_sequences.sh <trim_left_FW> <trim_left_RV> <trunc_left_FW> <trunc_left_RV>
Inputs:
- <trim_left_FW>: number of bases to be trimmed from 5' end of forward reads
- <trim_left_RV>: number of bases to be trimmed from 5' end of reverse reads
- <trunc_left_FW>: number of bases to truncate forward reads at (choose a value by looking at demux_summary.qzv file)
- <trunc_left_RV>: number of bases to be truncate reverse reads at (choose a value by looking at demux_summary.qzv file)
Outputs:
- <rep-seqs.qz*>: representative sequences (Amplicon Sequence Variants, ASVs)
- <table.qz*>: table with counts for each ASV
- <denoising-stats.qzv>: statistics describing denoising performed by DADA2
Assign_taxonomy.sh
Usage: Assign_taxonomy.sh <classifier>
Inputs:
- <classifier>: QIIME2 artifact (.qza) for Naive-Bayes classifier
Outputs:
- <taxonomy.qz*>: taxonomy assigned to each ASV
- <taxa-bar-plots.qzv>: taxonomy barplot describing samples composition at each taxonomic level
- <feature-table_*freq.tsv>: tables with the number or proportion of reads assigned to each taxa
Diversity_analyses.sh
Usage: Diversity_analyses.sh <sampling_depth>
Inputs:
- <sampling_depth>: minimum number of filtered fragments for a sample to be included in the analyses; the value should be chosen by looking at table.qzv file
Outputs:
- <core-metrics-results>: directory containing various beta diversity analyses
- <alpha-rarefaction.qzv>: rarefaction curve, computing the dependance between sequencing depths and various alpha diversity metrics
All .qzv and .qza artifacts can be visualized either importing them to QIIME2 View or with command:
source activate QIIME2_Illumina_env
qiime tools view <file.qz*>
The QIIME2_Illumina pipeline is composed of some wrapper scripts for QIIME2.
If this tool is useful for your work, please consider citing our manuscripts.
Matoute, A.; Maestri, S.; Saout, M.; Laghoe, L.; Simon, S.; Blanquart, H.; Hernandez Martinez, M.A.; Pierre Demar, M. Meat-Borne-Parasite: A Nanopore-Based Meta-Barcoding Work-Flow for Parasitic Microbiodiversity Assessment in the Wild Fauna of French Guiana. Curr. Issues Mol. Biol. 2024, 46, 3810-3821. https://doi.org/10.3390/cimb46050237
Klain, V., Maestri, S., & Bicca-Marques, J. C. (2024). Landscape structure influences the eukaryome of a folivorous-frugivorous primate. Studies on Neotropical Fauna and Environment, 1–8. https://doi.org/10.1080/01650521.2024.2423559
Please, refer to the following manuscript for further information.
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, and Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37: 852–857. https://doi.org/10.1038/s41587-019-0209-9