A NextFlow pipeline for Metagenomic Sequencing with Spiked Primer Enrichment
Prerequisites
Installation
git clone https://github.com/MaestSi/nf-msspe.git
cd nf-msspe
chmod 755 *
The nf-msspe pipeline requires you to open nf-msspe.conf configuration file and set the desired options. Then, you can run the pipeline using either docker or singularity environments just specifying a value for the -profile variable.
Usage:
nextflow -c nf-msspe.conf run nf-msspe.nf --fasta_file = "/path/to/file.fasta" --primers_file = "/path/to/primers.fasta" -profile docker
Mandatory argument:
-profile Configuration profile to use. Available: docker, singularity
Other mandatory arguments which may be specified in the nf-msspe.conf file
--fasta_file = "/path/to/file.fasta" Path to fasta file with genomes of interest
--primers_file = "/path/to/primers.fasta" Path to output fasta file with primers
--ovlp_window_size = 250 Size of windows overlaps
--search_window_size = 50 Search window size at the extremities of windows
--kmer_size = 13 Primers length
--num_acc_miss = 0 Number of accepted missed sequences, to reduce costs
--num_max_it = 1000 Maximum number of iterations
--scripts_dir = "/path/to/scripts/dir" Path to directory containing MSSPE.R script
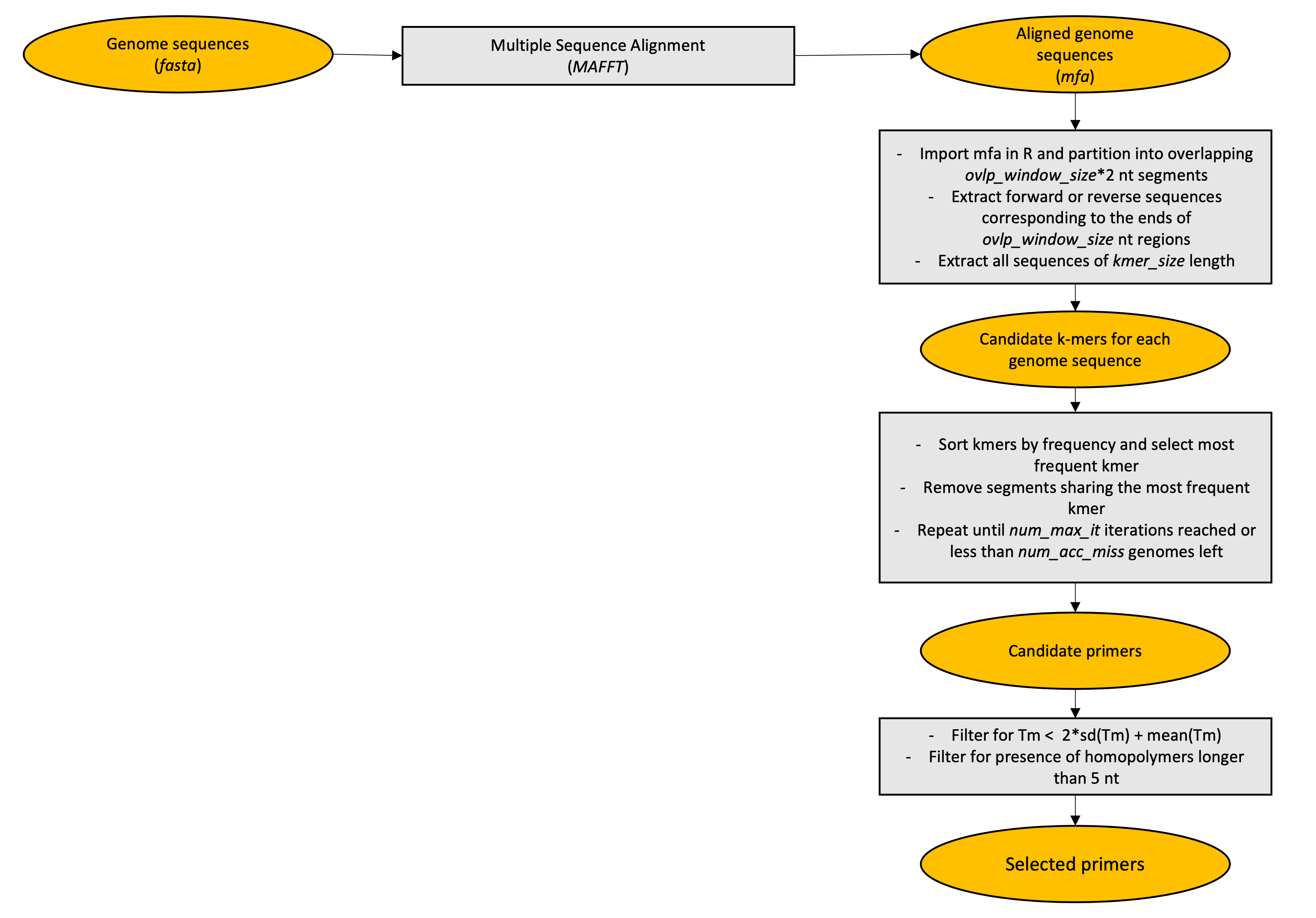
This pipeline is a NextFlow implementation of MSSPE pipeline described in Deng et al. paper, with minor adjustments.
Deng, X., Achari, A., Federman, S. et al. Metagenomic sequencing with spiked primer enrichment for viral diagnostics and genomic surveillance. Nat Microbiol 5, 443–454 (2020).