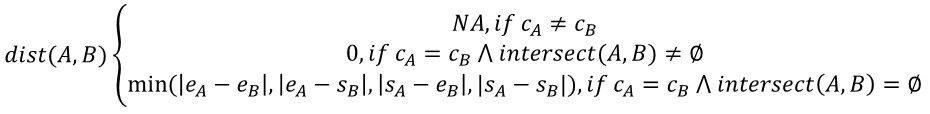This is a R/C++ (using Rcpp and RcppParallel) implementation of the distance function used by the popular GenomicRanges Bioconductor package. This tool is meant to be used when we wish to find the nearest neighbor in a set of reference or ranges R for each range in a query set Q and the corresponding distance (i.e. Calculate all |R| distances for each range in Q, then record minimum distance and index of minimum).
Calculating the resulting minimum distances and neighbors matrix can be very costly with the GenomicRanges functions when the query and reference sets are big. For a pair of ranges (most commonly genomic regions) A and B, each with integer properties c (sequence, most commonly the chromosome), s (start position) and e (end position), the block distance (as used by GenomicRanges v1.42.0) between them is defined as:
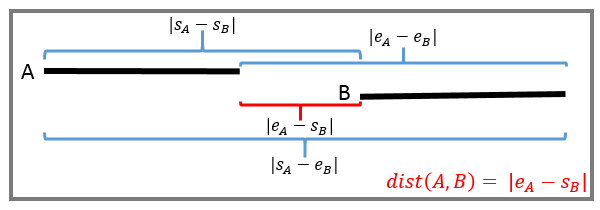
Where intersect(A, B) is a function that computes the set of overlapping positions between the two ranges. The third case of the equation when ranges have no overlapping positions is better understood with a visual example:
Note that a gap of 1 position in the last case is defined as a distance of 0 in GenomicRanges v1.42.0.
Two matrices, one for the query and one for the reference. Each row of the matrices represents a range. The matrices should have 3 columns denoting the properties of each range: sequence (most commonly the chromosome), start position in the sequence and end posiion in the sequence, in that order.
A matrix with as many rows as the query had. The first column denotes the distance to the nearest neighbor in the reference set and the second column the index of the reference range that is the nearest neighbor for the corresponding query range.
The file benchmark.r has a simulation where one can verify that this tool produces the same results as GenomicRanges with an average improvement from 2.5 seconds to 0.001 when processing a query of 500 ranges and a reference of 500 ranges. This improvement is observed even without using additional cores. The script also serves as a usage example.