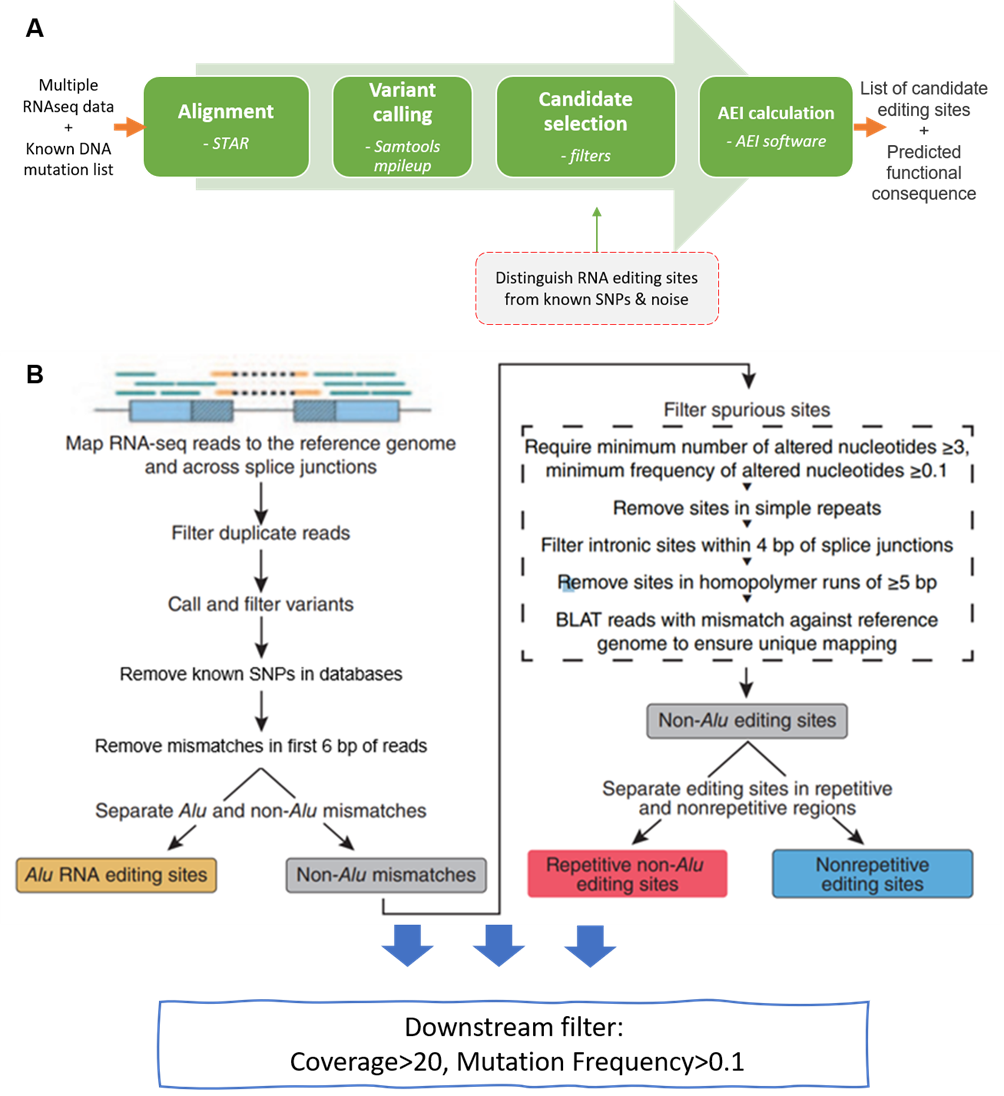
A shell pipeline for RNA-editing site identification from RNA-seq data only
RNA editing is a post-transcriptional modification to alter the sequence of RNA molecules while keeping the original DNA sequence unchanged. This is a shell pipeline used in linux server to extract the valid RNA-editing site from RNA-seq data. The overall pipeline is adopted from CSI NUS Portal and Ramaswami's pipeline (2013) with a series of filters to exclude the technical biases.
- reference_file_download.sh includes all reference files used in this pipeline.
- SRA_workflow_full.sh is the main pipeline that cover the entire analyzing and filtering process.
- perlCode repository keeps the specific filtering procedure that is written in perl document and was excuted in the overall workflow file.
The pipeline is applied in my undergraudate thesis: Investigating the role of ADAR-mediated RNA-editing in Cardiomyopathy.
I would like to acknowledge Prof Roger Foo's supervise, guidance, and direction suggestions on this project, Dr Willson Tan, and Dr LEE Chang Jie Mick's suggestions in bioinformatics pipeline establishment and improvement. Also, I would like to thank Prof Polly Chen and her postdoc Dr Omer An's support for providing CSI bioinformatic portal and their related pipeline documents, Prof Gokul Ramaswami for providing the original RNA-editing pipeline codes. Thanks to Professor Robert Young for being my link-supervisor, giving me frequent supervising on my project progress, and providing access and technical support for the use of the UoE Eddie Remote Server.