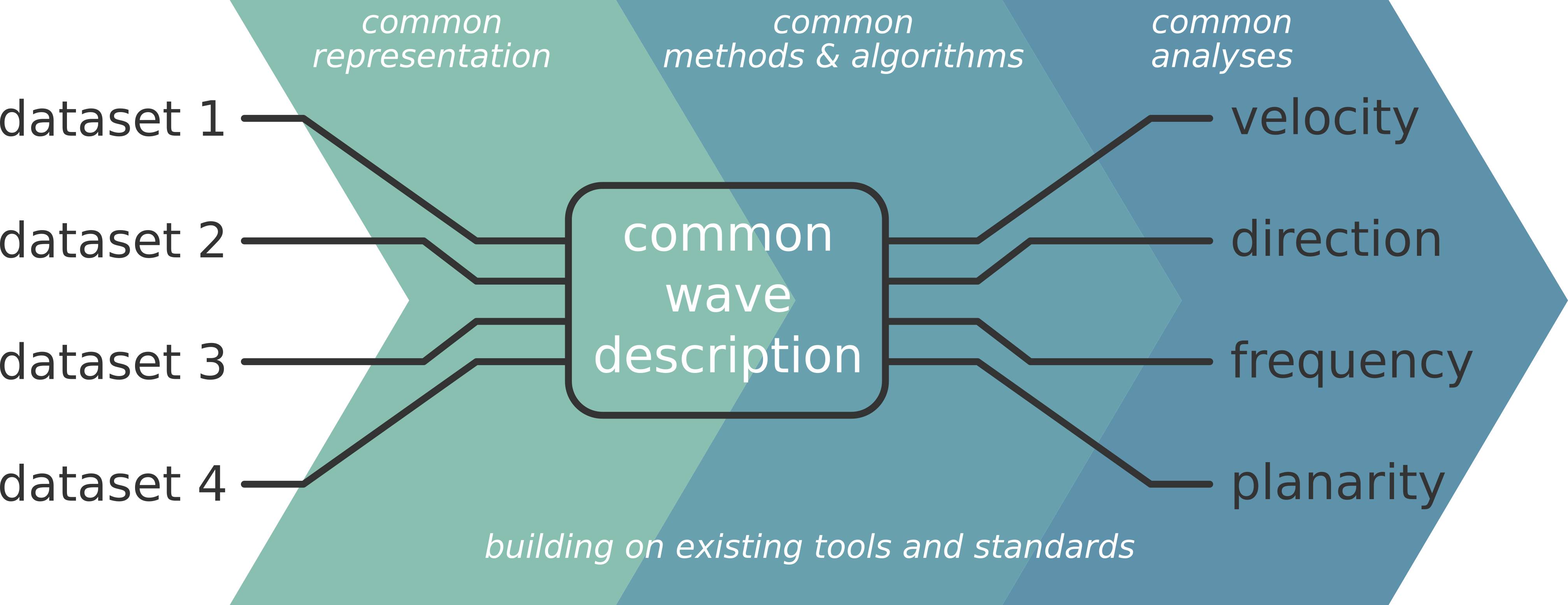
Cobrawap is an adaptable and reusable analysis pipeline for the multi-scale, multi-methodology analysis of cortical wave activity. The pipeline ingests data from heterogeneous sources of spatially organized neuronal activity, such as ECoG or calcium imaging recordings, as well as the outcome of numerical simulations. The pipeline returns statistical measures to quantify the dynamic wave-like activity patterns found in the data.
Documentation | Publication | Introductory video | Use-case demo on the EBRAINS Collab
Cobrawap brings together existing analysis methods, tools, and standards, and interfaces them for the analysis and characterization of cortical wave-like activity and UP/DOWN state detections. Cobrawap serves as a space to gather the various data types exhibiting wave-like activity and their various analysis approaches into the same pipeline. Besides generating easily reproducible and curated results, Cobrawap facilitates the rigorous comparison between datasets of different laboratories, studies and measurement modalities. Furthermore, Cobrawap can be used in the context of model validation and benchmarking of analysis methods. Cobrawap may also act as a template for implementing analysis pipelines in other contexts.
Cobrawap features...
- a user-friendly command line interface guiding the setup and usage
- a hierarchical and modular pipeline framework based on the Snakemake workflow management tool
- reusable method implementations (stages and blocks) for standalone applications or integration into workflows
- analysis methods for electrophysiological and optical data on the characterization of cortical wave activity and local oscillations
- visualization of the analysis steps and the intermediate results
- intermediate results curated with annotated metadata
For further developments and feature requests refer to the Github Issues.
To refer to the Cobrawap software package in publications, please use:
Cobrawap (doi:10.5281/zenodo.10198748; RRID:SCR_022966)
To cite a specific version of Cobrawap please see version-specific DOIs at:
doi:10.5281/zenodo.10198748
To cite Cobrawap, please use:
Gutzen, R., De Bonis, G., De Luca, C., Pastorelli, E., Capone, C., Allegra Mascaro, A. L., Resta, F., Manasanch, A., Pavone, F. S., Sanchez-Vives, M. V., Mattia, M., Grün, S., Paolucci, P. S., & Denker, M. (2024). Using a modular and adaptable analysis pipeline to compare slow cerebral rhythms across heterogeneous datasets. Cell Reports Methods, 4(1), 100681. https://doi.org/10.1016/j.crmeth.2023.100681
Cobrawap is open-source software and is licensed under the GNU General Public License v3.
Cobrawap is currently provided as a tool of the EBRAINS infrastructure and included in the EBRAINS-Italy initiative. Further details on funding and resources are in the Acknowledgments file in the doc folder.
The Cobrawap Core Team is in charge of defining the scientific address of the project and taking care of the continuous maintenance and development of the software. This collaborative endeavor is jointly carried by Forschungszentrum Jülich, Germany and Istituto Nazionale di Fisica Nucleare (INFN), Roma, Italy and currently includes:

|

|
|
|
The further Cobrawap Community includes people and partners that offer technical support for the integration of the software in a larger framework of interoperable tools, and offer scientific support for the development of the analysis methods and the tool's integration into broader research endeavors.
- Athena Research and Innovation Center, Greece
- Sofia Karvounari
- Eleni Mathioulaki
- Unité de Neurosciences, Neuroinformatics Group, CNRS, France
- Andrew Davison
- Istituto Nazionale di Fisica Nucleare (INFN), Italy
- Chiara De Luca
- Cristiano Capone
- Irene Bernava
- Alessandra Cardinale
- Institut d’Investigacions Biomediques August Pi i Sunyer (IDIBAPS), Barcelona, Spain
- Arnau Manasanch
- Miguel Dasilva
- Maria V. Sanchez-Vives
- European Laboratory for Non-Linear Spectroscopy (LENS), Firenze, Italy
- Anna Letizia Allegra Mascaro
- Francesco Resta
- Francesco S. Pavone
- Istituto Superiore di Sanità (ISS), Roma, Italy
- Maurizio Mattia
- University of Milano (UniMi), Italy
- Andrea Pigorini
- Thierry Nieus
- Gianluca Gaglioti
- Marcello Massimini
Cobrawap Partnering Projects:
Sleep Wave Analysis Visualization Engine (SWAVE): A data visualization tool that takes Cobrawap outputs and visualizes these to show dynamic wave-like activity patterns found in the data. Developed at Washington University in St. Louis, USA: https://github.com/cilantroxiao/SWAVE
The functionality offered by Cobrawap builds on existing software tools and services.
Neo improves interoperability between Python tools for analyzing, visualizing, and generating electrophysiology data by providing a common, shared data object model. The Neo data representation provides a hierarchical data and metadata description for a variety of data types including intracellular and extracellular electrophysiology, electrical data with support for multi-electrode, as well as optical recordings. Furthermore, it supports a wide range of neurophysiology file formats to facilitate reading data from most common recording devices.
The Electrophysiology Analysis Toolkit, Elephant, is an open-source Python library for analysis methods. It focuses on providing fast and reliable implementations for generic analysis functions for spike train data and time series recordings from electrodes. As community centered project, Elephant aims to serve as a common platform for analysis codes from different laboratories, and a consistent and homogeneous analysis framework.
The Neuroscience Information Exchange, NIX, format is an API and data format to store scientific data and metadata in a combined representation. Its structure is inspired by common types of neuroscience data, and it acts as one of the primary data formats for the Neo data object model.
Cobrawap was originally developed in the context the Human Brain Project, launched as a use-case initiated within the WaveScalES sub-project. Sleep is present in all animal species notwithstanding the risk associated with the disconnection from the environment (e.g. predation) and the reduction of time available for food search and reproduction. Indeed, it is well known that the human brains need healthy sleep, as chronic sleep deprivation reduces cognitive performances. The goal of the WaveScalES sub-project of the Human Brain Project was to unveil the underlying mechanisms of deep sleep, anesthesia and coma, the emergence toward wakefulness, and the link between sleep and learning, taking advantage of cortical slow wave activity (SWA) and investigating it with experimental data, analysis tools, modulation techniques, theoretical models, and simulations of such states and of the transition to wakefulness.