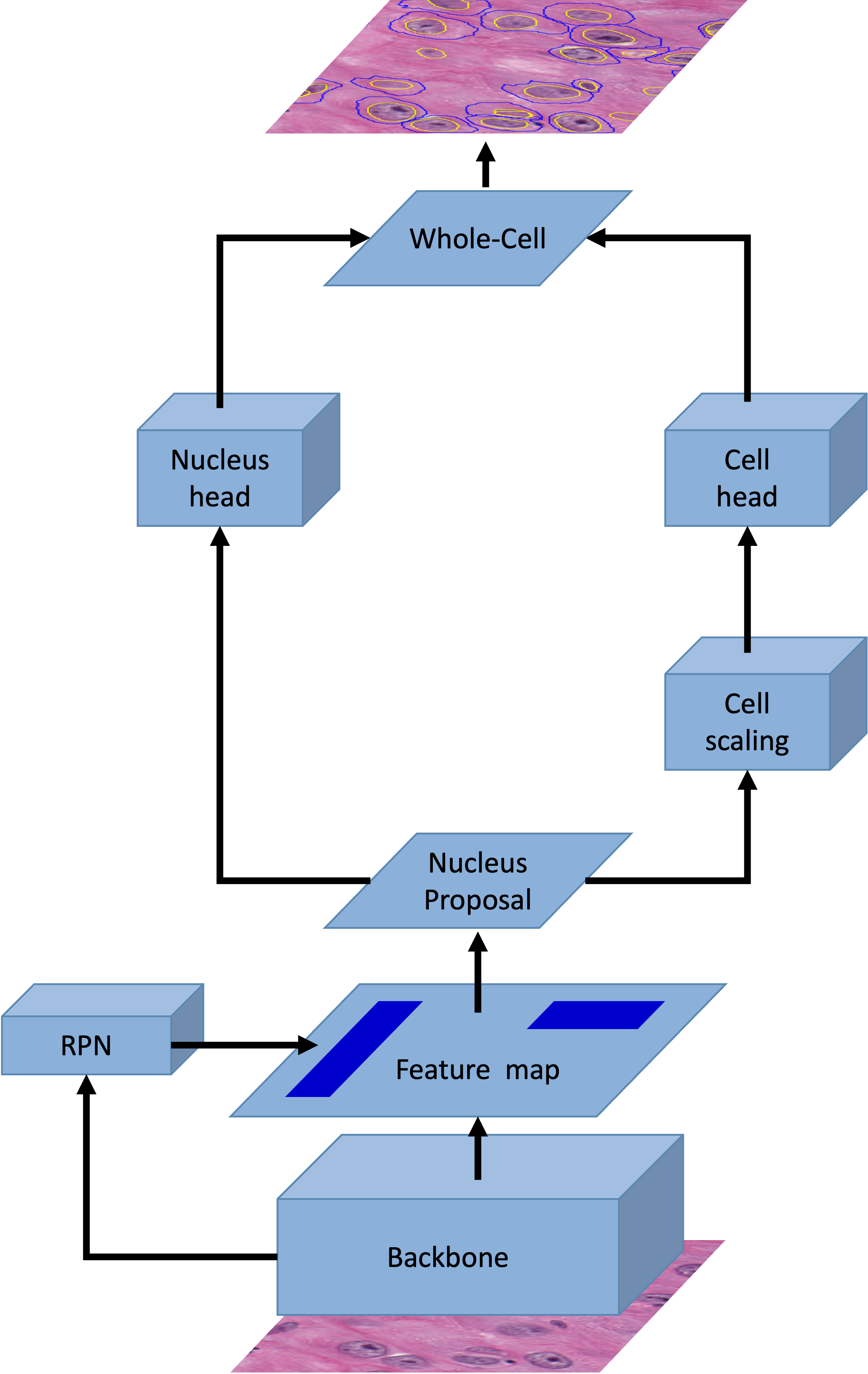
Cyto R-CNN and CytoNuke Dataset: Towards reliable whole-cell segmentation in bright-field histological images
This repository contains the Python implementation of Cyto R-CNN.
First, you will need to install CUDA on your machine. This code has been developed with Python 3.9 and CUDA 11.8.
conda create -n cytorcnn python=3.9conda activate cytorcnn- Install an appropriate version of torch, torchvision and CUDA.
pip install -r requirements.txtpip install 'git+https://github.com/facebookresearch/detectron2.git'pip install -e .
The CytoNuke dataset can be downloaded from Zenodo.
You can download a pretrained for the CytoNuke dataset on Google Drive
## Prediction
from cytorcnn import CytoRCNN, utilities
weights_path = "/.../model.pth"
image_path = "/.../image.png"
cytorcnn = CytoRCNN(weights_path)
cytorcnn.predict(image_path)
## Training
train_images = "/.../train/images"
train_coco = "/.../train/coco.json"
train_dataset = Dataset(train_images, train_coco)
val_images = "/.../val/images"
val_coco = "/.../val/coco.json"
val_dataset = Dataset(val_images, val_coco)
# Optional: Register run in WeightsAndBiases
wandb.init(
project="CytoRCNN"
)
cytorcnn = CytoRCNN()
cytorcnn.train(train_dataset, val_dataset)@article{raufeisen2024,
title={Cyto R-CNN and CytoNuke Dataset: Towards reliable whole-cell segmentation in bright-field histological images},
journal = {Computer Methods and Programs in Biomedicine},
volume = {252},
pages = {108215},
year={2024},
issn = {0169-2607},
author={Johannes Raufeisen and Kunpeng Xie and Fabian Hörst and Till Braunschweig and Jianning Li and Jens Kleesiek and Rainer Röhrig and Jan Egger and Bastian Leibe and Frank Hölzle and Alexander Hermans and Behrus Puladi},
doi = {https://doi.org/10.1016/j.cmpb.2024.108215},
url = {https://www.sciencedirect.com/science/article/pii/S0169260724002104},
}