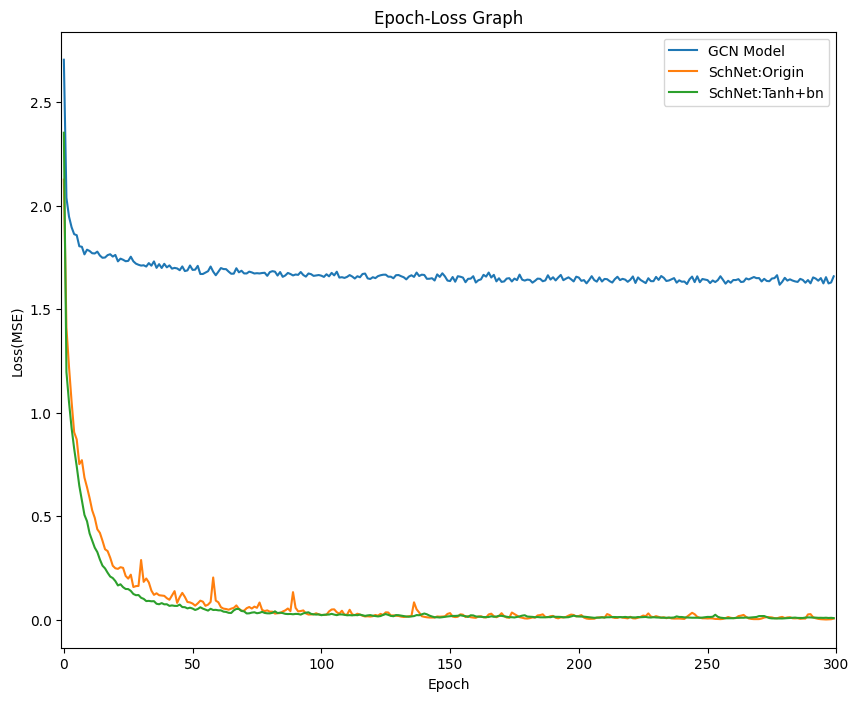
Given amino acids molecule datasets, we want to predict the dipole moment of other amino acids.
In this project, we used modified SchNet model to improve the overall performance and tested out with GCN model for comparison.
Data Information:
- Train Data: 10984
- Test Data: 1217
- Contains: x, edge_index, edge_attr, y, pos, batch, ptr
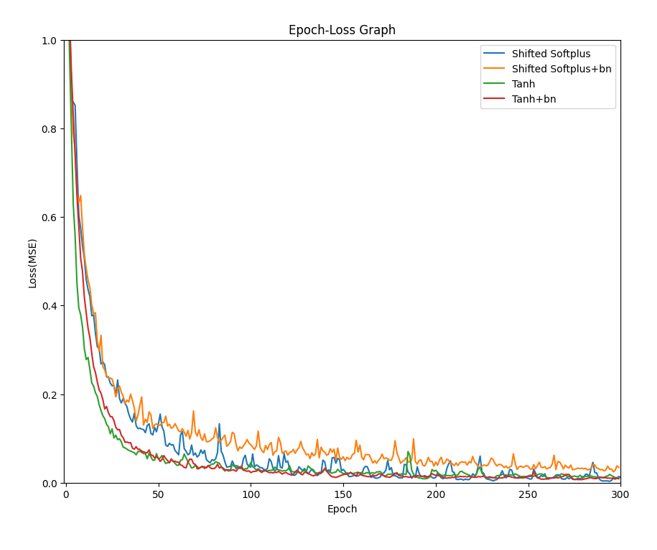
What we modified:
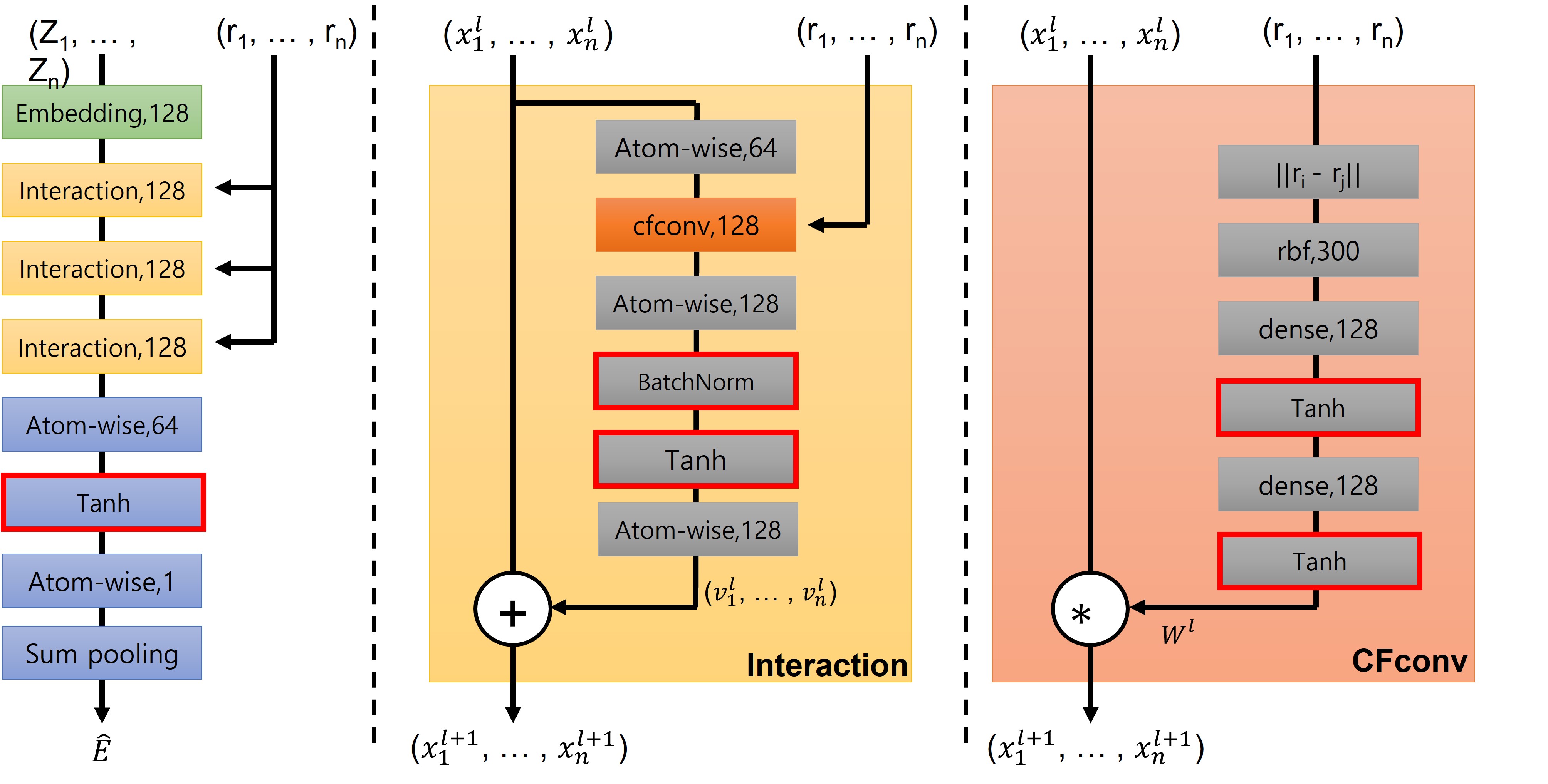
- Added batch layer normalization
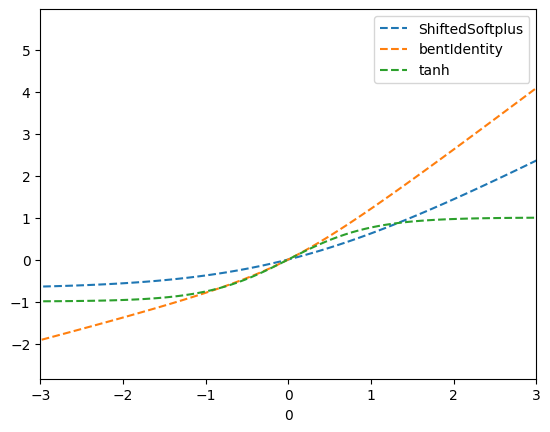
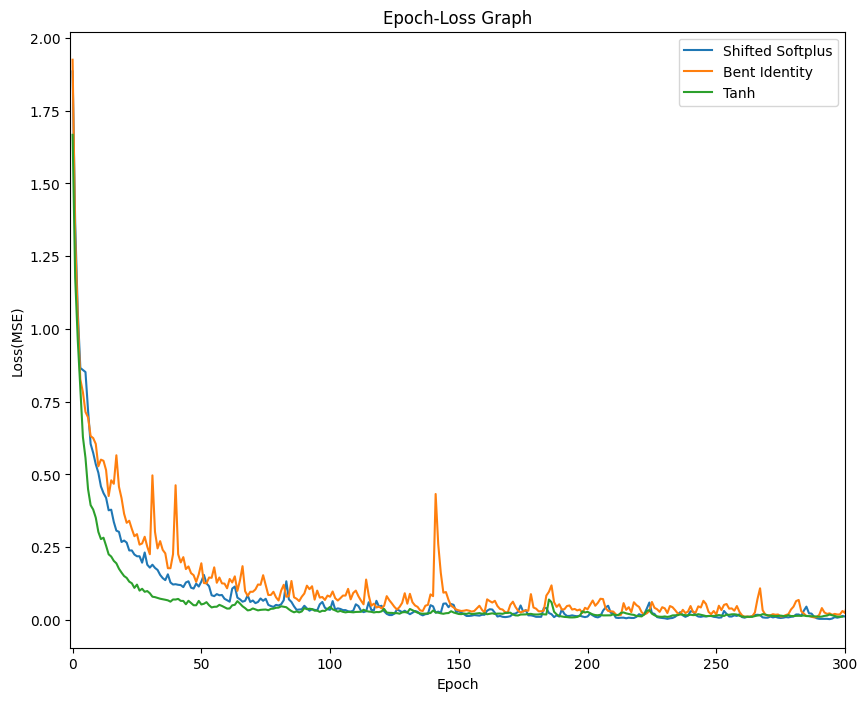
- Changed activation function to Tanh
- Reduced the number of interactions to 4
- Add bias to the linear layer
Training Configurations:
- Batch Size: 64
- Trainset Shuffled
- Learning Rate: 0.0008
- Weight Decay: 0
- Adam Optimizer
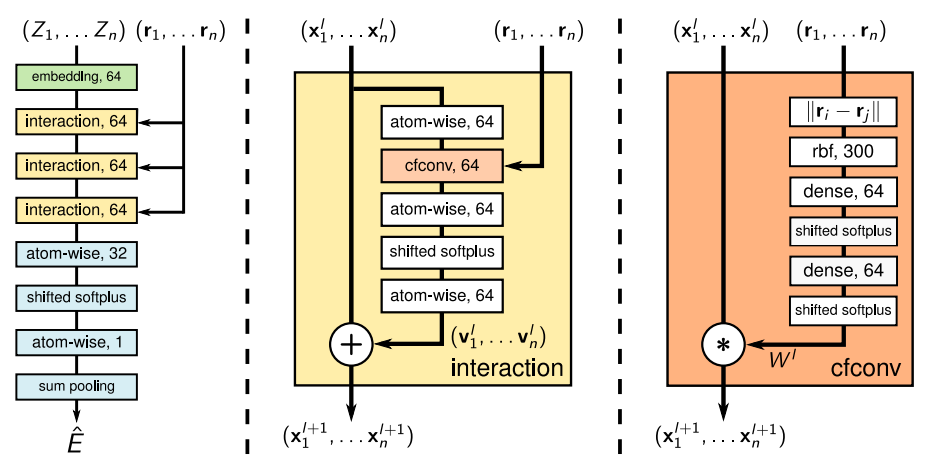
Original SchNet Architecture
Modified SchNet Architecture
1. Pytorch
2. Pytorch Geometric
3. Rdkitimport os
import os.path as osp
import warnings
import torch
import pandas as pd
import numpy as np
import math
import torch_geometric.transforms as T
import torch.nn as nn
import torch.nn.functional as F
from tqdm import tqdm
from glob import glob
from rdkit import Chem, RDLogger
from torch_geometric.data import Data
from torch_geometric.loader import DataLoader
from torch_geometric.nn.conv import MessagePassing
from torch_geometric.nn import global_mean_pool as gap, global_max_pool as gmp
from torch_geometric.nn import Linear
from math import pi as PI
from typing import Callable, Optional, Tuple
from torch import Tensor
from torch.nn import Embedding, Linear, ModuleList, Sequential
from torch_geometric.data import Dataset, download_url, extract_zip
from torch_geometric.data.makedirs import makedirs
from torch_geometric.nn import MessagePassing, SumAggregation, radius_graph
from torch_geometric.nn.resolver import aggregation_resolver as aggr_resolver
from torch_geometric.typing import OptTensor# allowable multiple choice node and edge features
allowable_features = {
'possible_atomic_num_list' : list(range(1, 119)) + ['misc'],
'possible_bond_type_list' : [
'SINGLE',
'DOUBLE',
'TRIPLE',
'AROMATIC',
'misc'
],
}
def safe_index(l, e):
"""
Return index of element e in list l. If e is not present, return the last index
"""
try:
return l.index(e)
except:
return len(l) - 1
def atom_to_feature_vector(atom):
"""
Converts rdkit atom object to feature list of indices
:param mol: rdkit atom object
:return: list
"""
atom_feature = [
safe_index(allowable_features['possible_atomic_num_list'], atom.GetAtomicNum()),
]
return atom_feature
def bond_to_feature_vector(bond):
"""
Converts rdkit bond object to feature list of indices
:param mol: rdkit bond object
:return: list
"""
bond_feature = [
safe_index(allowable_features['possible_bond_type_list'], str(bond.GetBondType())),
]
return bond_feature
def from_mol(mol_file, y=None, smiles=None):
r"""Converts a Molecule data to a :class:`torch_geometric.data.Data`
instance.
Args:
y(list, optional): target value
mol_file (string, optional): The Mol filename string.
"""
RDLogger.DisableLog('rdApp.*')
if y is not None:
y = torch.tensor(y, dtype=torch.float)
mol = Chem.SDMolSupplier(mol_file, removeHs=False,
sanitize=False)
mol = mol[0]
xs = []
xc = []
for i, atom in enumerate(mol.GetAtoms()):
positions = mol.GetConformer().GetAtomPosition(i)
x = atom_to_feature_vector(atom)
xc.append([positions.x, positions.y, positions.z])
xs.append(x)
x = torch.tensor(xs, dtype=torch.float).view(-1, 1)
xc = torch.tensor(xc, dtype=torch.float).view(-1, 3)
edge_indices, edge_attrs = [], []
for bond in mol.GetBonds():
i = bond.GetBeginAtomIdx()
j = bond.GetEndAtomIdx()
e = bond_to_feature_vector(bond)
edge_indices += [[i, j], [j, i]]
edge_attrs += [e, e]
edge_index = torch.tensor(edge_indices)
edge_index = edge_index.t().to(torch.long).view(2, -1)
edge_attr = torch.tensor(edge_attrs, dtype=torch.float).view(-1, 1)
if edge_index.numel() > 0: # Sort indices.
perm = (edge_index[0] * x.size(0) + edge_index[1]).argsort()
edge_index, edge_attr = edge_index[:, perm], edge_attr[perm]
return Data(x=x, pos=xc, y=y, edge_index=edge_index, edge_attr=edge_attr, smiles=smiles)train = pd.read_csv(r"./train.csv")
test = pd.read_csv(r"./test.csv")
# ============== training data ================== #
for idx in tqdm(train.index):
y = [train["mu"][idx]]
i = train["Id"][idx]
data = from_mol(fr"./mol/train/{i}.mol", y=y)
torch.save(data,fr"./data/train/{i}.pt")
# ============== test data ================== #
for idx in tqdm(test.index):
i = test["Id"][idx]
data = from_mol(fr"./mol/test/{i}.mol")
torch.save(data, fr"./data/test/{i}.pt")
train = pd.read_csv(r"./train.csv", index_col=0)
test = pd.read_csv(r"./test.csv", index_col=0)
train_num_nodes_list = list()
test_num_nodes_list = list()
train_list = list()
test_list = list()
for idx in tqdm(train.index):
d = torch.load(fr"./data/train/{idx}.pt")
train_list.append(d)
train_num_nodes_list.append(d.num_nodes)
for idx in tqdm(test.index):
d = torch.load(fr"./data/test/{idx}.pt")
test_list.append(d)
test_num_nodes_list.append(d.num_nodes)class SchNet(torch.nn.Module):
url = 'http://www.quantum-machine.org/datasets/trained_schnet_models.zip'
def __init__(
self,
hidden_channels: int = 128,
num_filters: int = 128,
num_interactions: int = 4,
num_gaussians: int = 100, # 커지면 빠른 학습
cutoff: float = 10.0,
interaction_graph: Optional[Callable] = None,
max_num_neighbors: int = 32,
readout: str = 'mean',
dipole: bool = False,
mean: Optional[float] = None,
std: Optional[float] = None,
atomref: OptTensor = None,
):
super().__init__()
self.hidden_channels = hidden_channels
self.num_filters = num_filters
self.num_interactions = num_interactions
self.num_gaussians = num_gaussians
self.cutoff = cutoff
self.dipole = dipole
self.sum_aggr = SumAggregation()
self.readout = aggr_resolver('sum' if self.dipole else readout)
self.mean = mean
self.std = std
self.scale = None
self.embedding = Embedding(100, hidden_channels, padding_idx=0)
if interaction_graph is not None:
self.interaction_graph = interaction_graph
else:
self.interaction_graph = RadiusInteractionGraph(
cutoff, max_num_neighbors)
self.distance_expansion = GaussianSmearing(0.0, cutoff, num_gaussians)
self.interactions = ModuleList() ## 들어갈 블럭을 만듬
for _ in range(num_interactions):
block = InteractionBlock(hidden_channels, num_gaussians,
num_filters, cutoff)
self.interactions.append(block)
self.lin1 = Linear(hidden_channels, hidden_channels // 2)
self.act = nn.Tanh()
self.lin2 = Linear(hidden_channels // 2, 1)
self.bn =nn.BatchNorm1d(hidden_channels // 2)
self.register_buffer('initial_atomref', atomref)
self.reset_parameters()
def reset_parameters(self):
self.embedding.reset_parameters()
for interaction in self.interactions:
interaction.reset_parameters()
torch.nn.init.xavier_uniform_(self.lin1.weight)
self.lin1.bias.data.fill_(0)
torch.nn.init.xavier_uniform_(self.lin2.weight)
self.lin2.bias.data.fill_(0)
def forward(self, z: Tensor, pos: Tensor,
batch: OptTensor = None) -> Tensor:
batch = torch.zeros_like(z) if batch is None else batch
h = self.embedding(z)
edge_index, edge_weight = self.interaction_graph(pos, batch)
edge_attr = self.distance_expansion(edge_weight) # RDF값
for interaction in self.interactions:
h = h + interaction(h, edge_index, edge_weight, edge_attr)
h = self.lin1(h)
h = self.bn(h)
h = self.act(h)
h = self.lin2(h)
out = self.readout(h, batch, dim=0)
return out
def __repr__(self) -> str:
return (f'{self.__class__.__name__}('
f'hidden_channels={self.hidden_channels}, '
f'num_filters={self.num_filters}, '
f'num_interactions={self.num_interactions}, '
f'num_gaussians={self.num_gaussians}, '
f'cutoff={self.cutoff})')
class RadiusInteractionGraph(torch.nn.Module):
def __init__(self, cutoff: float = 10.0, max_num_neighbors: int = 32):
super().__init__()
self.cutoff = cutoff
self.max_num_neighbors = max_num_neighbors
def forward(self, pos: Tensor, batch: Tensor) -> Tuple[Tensor, Tensor]:
edge_index = radius_graph(pos, r=self.cutoff, batch=batch,
max_num_neighbors=self.max_num_neighbors)
row, col = edge_index
edge_weight = (pos[row] - pos[col]).norm(dim=-1)
return edge_index, edge_weight
class InteractionBlock(torch.nn.Module):
def __init__(self, hidden_channels: int, num_gaussians: int,
num_filters: int, cutoff: float):
super().__init__()
self.mlp = Sequential(
Linear(num_gaussians, num_filters),
nn.Tanh(),
Linear(num_filters, num_filters),
)
self.conv = CFConv(hidden_channels, hidden_channels, num_filters,
self.mlp, cutoff)
self.act =nn.Tanh()
self.lin = Linear(hidden_channels, hidden_channels)
self.reset_parameters()
def reset_parameters(self):
torch.nn.init.xavier_uniform_(self.mlp[0].weight)
self.mlp[0].bias.data.fill_(0)
torch.nn.init.xavier_uniform_(self.mlp[2].weight)
self.mlp[2].bias.data.fill_(0)
self.conv.reset_parameters()
torch.nn.init.xavier_uniform_(self.lin.weight)
self.lin.bias.data.fill_(0)
def forward(self, x: Tensor, edge_index: Tensor, edge_weight: Tensor,
edge_attr: Tensor) -> Tensor:
x = self.conv(x, edge_index, edge_weight, edge_attr)
x = self.act(x)
x = self.lin(x)
return x
class CFConv(MessagePassing):
def __init__(self, in_channels: int, out_channels: int, num_filters: int,
nn: Sequential, cutoff: float):
super().__init__(aggr='mean')
self.lin1 = Linear(in_channels, num_filters, bias=True)
self.lin2 = Linear(num_filters, out_channels)
self.nn = nn
self.cutoff = cutoff
self.reset_parameters()
def reset_parameters(self):
torch.nn.init.xavier_uniform_(self.lin1.weight)
torch.nn.init.xavier_uniform_(self.lin2.weight)
self.lin2.bias.data.fill_(0)
def forward(self, x: Tensor, edge_index: Tensor, edge_weight: Tensor,
edge_attr: Tensor) -> Tensor:
C = 0.5 * (torch.cos(edge_weight * PI / self.cutoff) + 1.0) # 웨이트로 계산과정
W = self.nn(edge_attr) * C.view(-1, 1) # 웨이트 계산 웨이트는 pos과 분자의 dimension을 계산
x = self.lin1(x)#
x = self.propagate(edge_index, x=x, W=W)
x = self.lin2(x)
return x
def message(self, x_j: Tensor, W: Tensor) -> Tensor:
return x_j * W
class GaussianSmearing(torch.nn.Module):##RDF용
def __init__(self, start: float = 0.0, stop: float = 5.0,
num_gaussians: int = 50):
super().__init__()
offset = torch.linspace(start, stop, num_gaussians)
self.coeff = -0.5 / (offset[1] - offset[0]).item()**2
self.register_buffer('offset', offset)
def forward(self, dist: Tensor) -> Tensor:
dist = dist.view(-1, 1) - self.offset.view(1, -1) # edge_weight = (pos[row] - pos[col]).norm(dim=-1). 분자사이 거리를 동해 회전 불변성을 얻는다. 에시)매트릭스 회전 원래값
return torch.exp(self.coeff * torch.pow(dist, 2)) # RDF 뉴럴 네트워크가 linear해지는 것을 방지한다. linear해지면 트랜딩 학습에 어려움
# num_gaussians증가 |self.coeff| 증가 더 뾰족 RDF
class ShiftedSoftplus(torch.nn.Module):
def __init__(self):
super().__init__()
self.shift = torch.log(torch.tensor(2.0)).item()
def forward(self, x: Tensor) -> Tensor:
return F.softplus(x) - self.shift
class bentIdentity(torch.nn.Module):
def __init__(self):
super().__init__()
def forward(self, x: Tensor) -> Tensor:
return ((torch.sqrt(x**2 + 1) - 1)/2) + xclass Model(torch.nn.Module):
def __init__(self):
super().__init__()
self.layer = SchNet()
def forward(self, data):
x, edge_index, batch, edge_attr, pos = data.x, data.edge_index, data.batch, data.edge_attr, data.pos
x = x.long()
x = (self.layer(x.squeeze(1), pos,batch))
return xtrainset = DataLoader(train_list, batch_size = 64, shuffle = True)
testset = DataLoader(test_list, batch_size = 64, shuffle = False)
device = torch.device('cuda')
model = Model().to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=0.0008, weight_decay=0e-4)
model.train()
for epoch in range(1000):
lossSum,count=0,0
for batch in trainset:
batch.to(device)
optimizer.zero_grad()
out = model(batch)
loss = torch.nn.MSELoss()(out.squeeze(1), batch.y)
loss.backward()
optimizer.step()
lossSum+=loss
count+=1
total_loss = lossSum/count
print("Epoch: {}, Loss: {:.6f}".format(epoch, total_loss))
if total_loss<=0.0018:
break
print('Training process has finished!')output = list()
model.eval()
for batch in testset:
batch.to(device)
predicted = model(batch)
for value in predicted:
output.append(value.item())submission = pd.DataFrame(output)
name=[]
for i in range(len(output)):
name.append('test_{0}'.format(i))
submission.index=name
submission.head(3)
submission.columns=['predicted']
submission.to_csv('./our_Submission.csv')
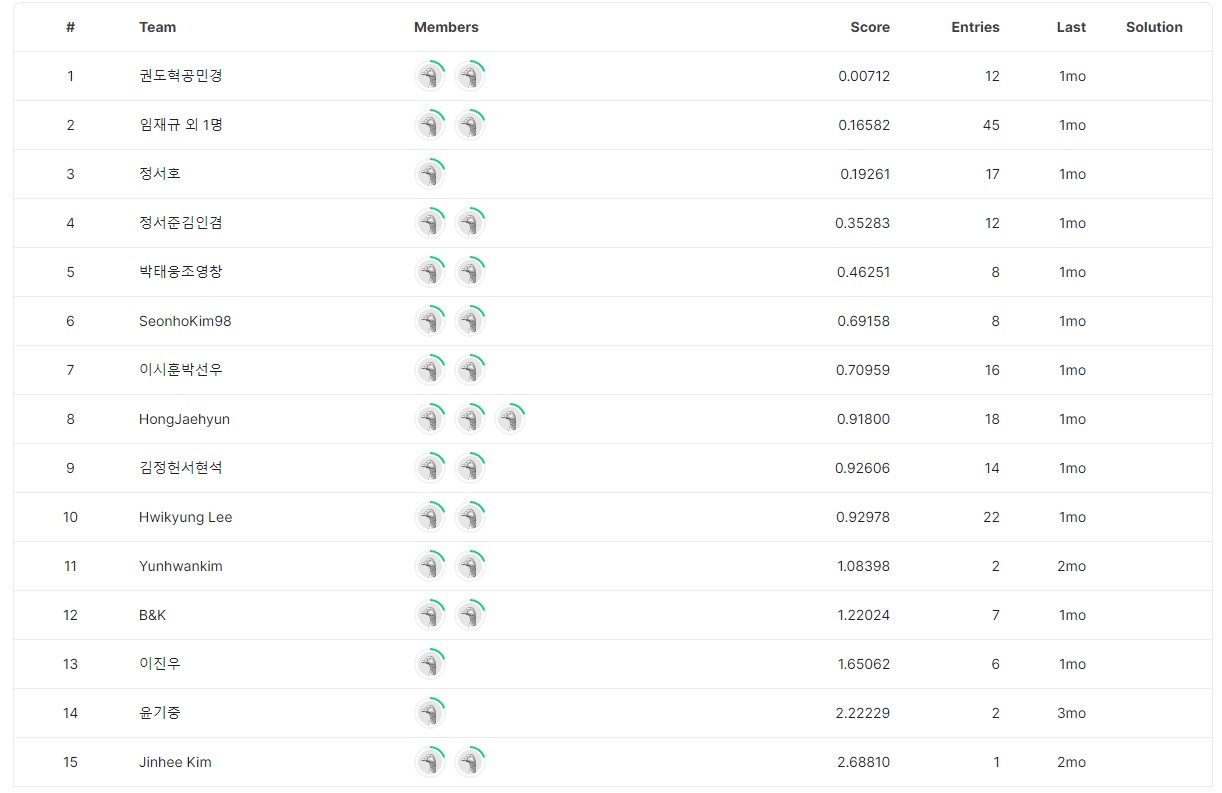
submission.head(3)Our team ranked 2nd
1. Clone this repository
2. Unzip mol file
3. Run the code in order from procedure 1 to 6