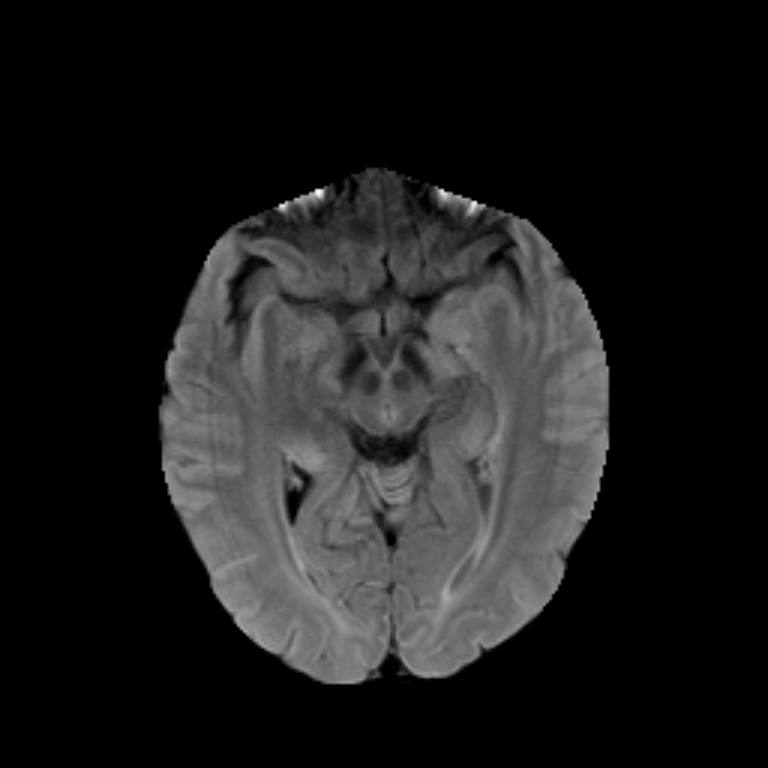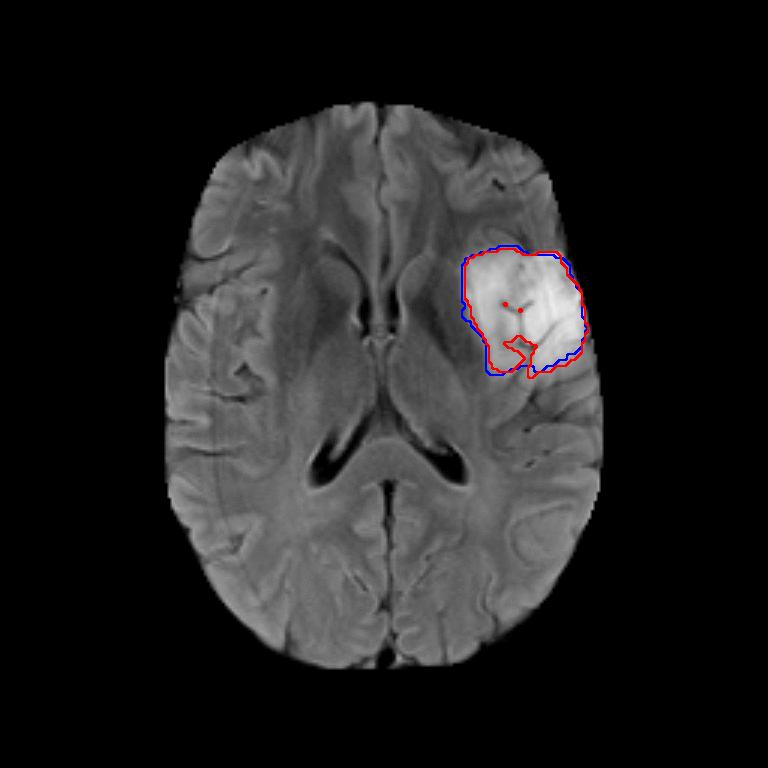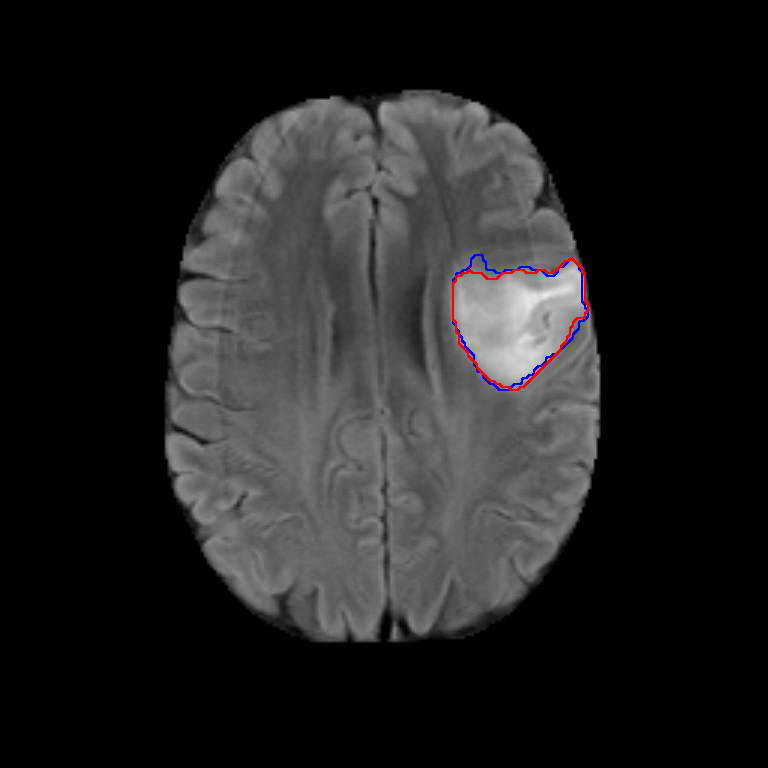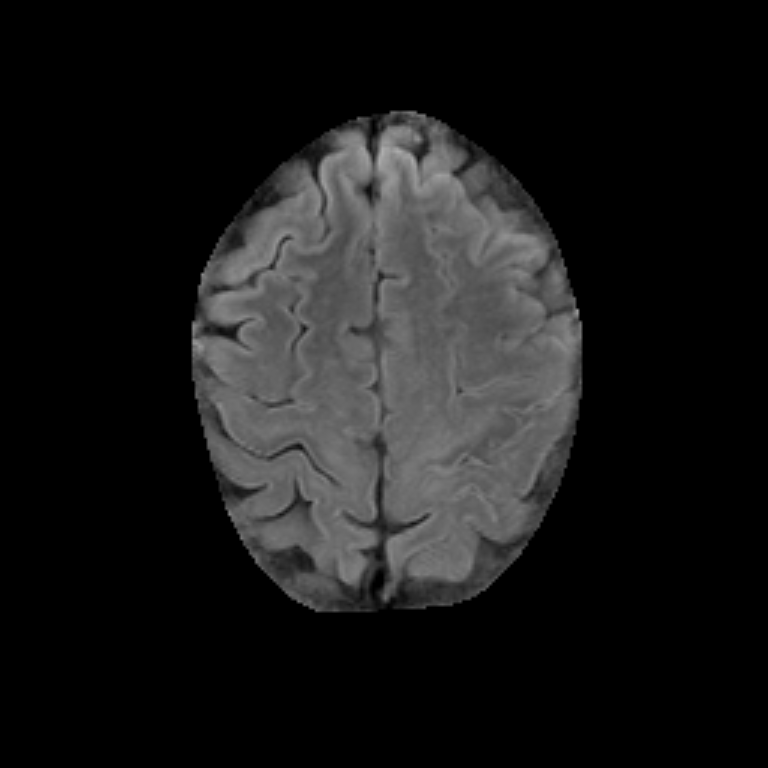
The dataset used for development was obtained from The Cancer Imaging Archive (TCIA) and involved 110 cases of lower-grade glioma patients. Registers brain MR images with manual FLAIR abnormality segmentation masks are published as a Kaggle Dataset lgg-mri-segmentation.
U-net:
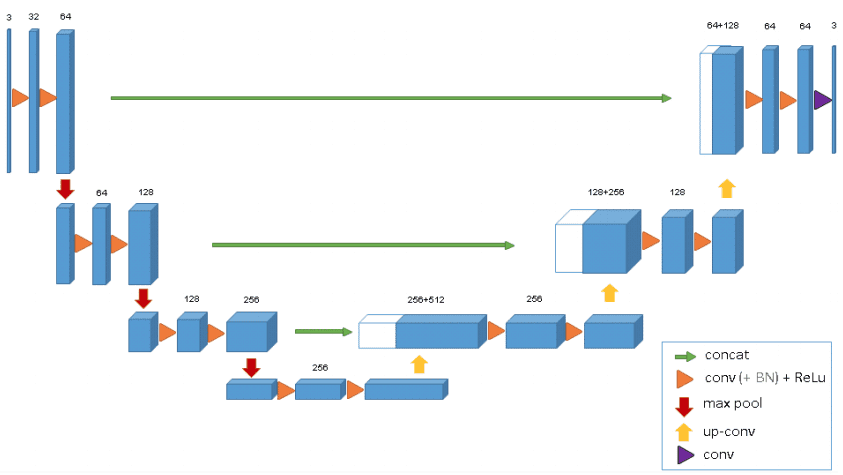
U-net from scratch has been written.
Key aspects of U-Net:
-
Convolution Layers: Convolution operation are used to learn information from images which then can be used as features for machine learning problems.
-
Down Sampling: Sequence of convolution combined with max pooling results in down sampling. In down sampling, size of the image is reduced which means we can observe larger portion of image in a single convolution operation. Down sampling is a good approach for identifying what is present in the image but for identifying where the object is we need to use upsampling.
-
Up Sampling: It is just opposite of down sampling. We go from low resolution to high resolution. For up sampling U-Net uses transposed covolution which is achieved by taking transpose of filter kernels and reversing the process of convolution.
Following picture gives a clear picture of What a U-Net is.
Reustls:
Dice Score on the test data: 0.9027