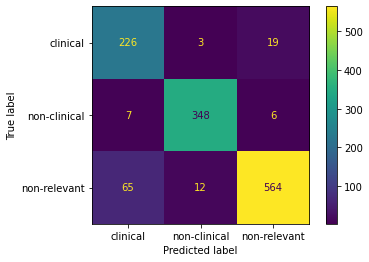
This component performs multiclass classification of abstracts to determine if they are relevant (either clinical or non-clinical studies) or not relevant to the field of biomaterials. We used the Transformers, the state-of-the-art of NLP, to train the DEBBIE_BioBERT model capable of detecting relevant biomaterials abstracts.
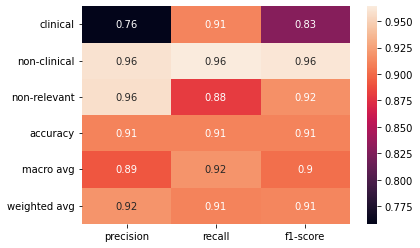
The DEBBIE_BioBERT model text-classification obtains an presicion of 0.92, a recall of 0.91 and an F1-score of 0.91. To achieve this result, we performed benchmarking between different pretrained models on biomedical data. The fine-tuning of the BioBERT (Bidirectional Encoder Representations from Transformers for Biomedical Text Mining) pretrained model obtained the best result. BioBERT is a domain-specific language representation model pre-trained on large-scale biomedical corpora. We made use of the BioBERT version pre-trained with PubMed.
This component was developed in Python leveraging the HuggingFace ecosystem for the development of transformer models https://huggingface.co/.
DEBBIE_BioBERT is available at https://huggingface.co/javicorvi/DEBBIE_BioBERT/
The DEBBIE_BioBERT has been fine-tuned with 3 collections:
- gold standard biomaterials non-clinical collection
- gold standard biomaterials clinical collection
- background set.
The folder classifier-dataset contains the test set that was used for validate the classifier.
The best model was selected after a benchmarking on different pre-trained biomedical models: BioBert, Bio_ClinicalBERT and BioDistilBERT-uncased.
The DEBBIE Abstract Classifier older version 3.00 was a logistic regression model with stochastic gradient descent optimization (SGDClassifier) using term frequency times inverse document frequency (TF-IDF). The old DEBBIE SGDClassifier model achieved an presicion of 0.90, a recall of 0.88 and an F1-score of 0.89. The new DEBBIE_BioBERT model achieved an presicion of 0.92, a recall of 0.91 and an F1-score of 0.91.
The performance results can be reproducible through the file debbie_abstract_classifier_performance.ypinb (jypiter notebook).
projectdebbie/classifier:4.0.0
python3 debbie_trained_classifier.py -i /in -o /out
#or in with docker
docker run --rm -u $UID -v ${PWD}/input_output:/in:ro -v ${PWD}/nlp_preprocessing_output:/out:rw projectdebbie/classifier:version python3 /usr/src/app/debbie_trained_classifier.py -i /in -o /out -w /usr/src/app
Parameters:
-i input folder with plain text abstracts
-o output folder with relevant biomaterials abstracts in plain text
-w work folder
- scikit-learn library
- Docker - Docker Containers
- Javier Corvi - Osnat Hakimi
This project is licensed under the GNU GENERAL PUBLIC LICENSE Version 3 - see the LICENSE file for details
 This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 751277
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 751277