The unofficial implementation of TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation on Pytorch
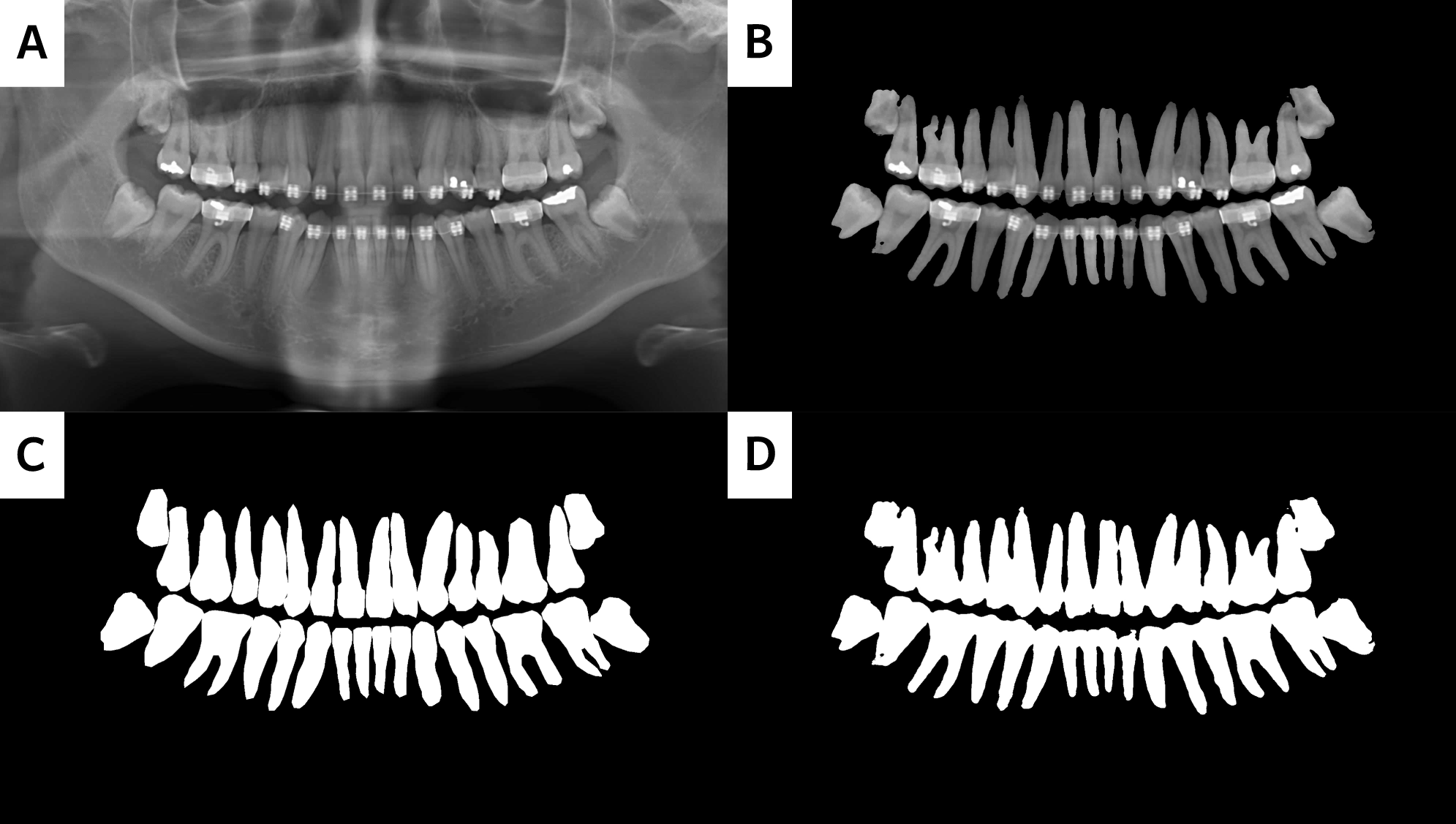 Output of my implementation. (A) Original X-Ray Image; (B) Merged Image of the Predicted Segmentation Map and Original X-Ray; (C) Ground Truth; (D) Predicted Segmentation Map
Output of my implementation. (A) Original X-Ray Image; (B) Merged Image of the Predicted Segmentation Map and Original X-Ray; (C) Ground Truth; (D) Predicted Segmentation Map
- On various medical image segmentation tasks, the ushaped architecture, also known as U-Net, has become the de-facto standard and achieved tremendous success. However, due to the intrinsic locality of convolution operations, U-Net generally demonstrates limitations in explicitly modeling long-range dependency. [1]
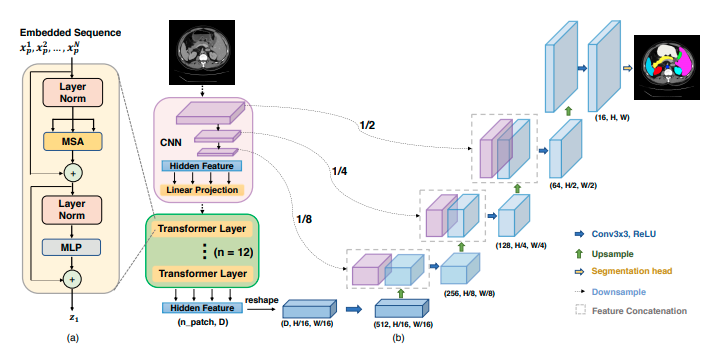
- TransUNet employs a hybrid CNN-Transformer architecture to leverage both detailed high-resolution spatial information from CNN features and the global context encoded by Transformers. [1]
TransUNet Architecture Figure from Official Paper
- Python 3.6+
pip install -r requirements.txt
- UFBA_UESC_DENTAL_IMAGES[2] dataset was used for training.
- Dataset can be accessed by request[3].
- Training process can be started with following command.
python main.py --mode train --model_path ./path/to/model --train_path ./path/to/trainset --test_path ./path/to/testset
- After model is trained, inference can be run with following command.
python main.py --mode inference --model_path ./path/to/model --image_path ./path/to/image