Code for the paper "Modelling protein complexes with crosslinking mass spectrometry and deep learning". We extend AlphaLink to protein complexes. AlphaLink2 is based on Uni-Fold and integrates crosslinking MS data directly into Uni-Fold. The current networks were trained with simulated SDA data (25 Å Cα-Cα).
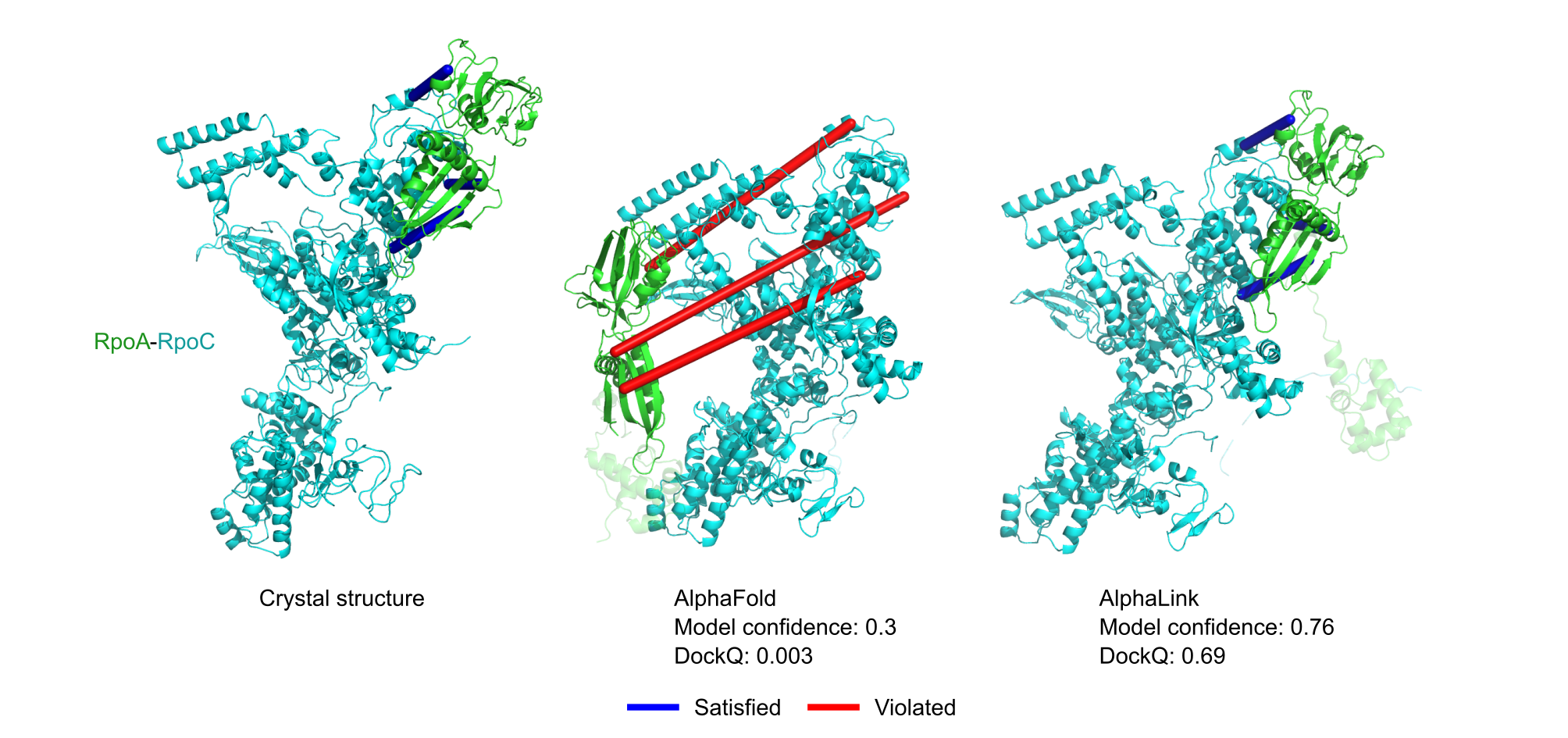
Figure 1. Prediction of RpoA-RpoC with real DSSO crosslinking MS data. Satisfied crosslinks are shown in blue (< 30 Å Cα-Cα) and violated crosslinks in red (> 30 Å Cα-Cα).The AlphaLink2 ColabFold can be found here.
AlphaLink takes as input a python dictionary of dictionaries with a list of crosslinked residue pairs with a false-discovery rate (FDR). That is, for inter-protein crosslinks A->B 1,50 and 30,80 and an FDR=20%, the input would look as follows:
In [6]: crosslinks
Out[6]: {'A': {'B': [(1, 50, 0.2), (30, 80, 0.2)]}}
Intra-protein crosslinks would go from A -> A
In [6]: crosslinks
Out[6]: {'A': {'A': [(5, 20, 0.2)]}}
The dictionaries are 0-indexed, i.e., residues start from 0.
You can create the dictionaries with the generate_crosslink_pickle.py script by running
python generate_crosslink_pickle.py --csv crosslinks.csv --output crosslinks.pkl.gz
The crosslinks CSV has the following format (residues are 1-indexed).
residueFrom chain1 residueTo chain2 FDR
Example:
1 A 50 B 0.2
5 A 5 A 0.1
The chain IDs A..Z+ designate all chains in the FASTA file, enumerated by order of appearance. That is, the first chain gets the identifier A, the second chain the identifier B and so on. After feature generation, the chain assignment can be found in the output folder in the file "chain_id_map.json" and the final composition in the file "chains.txt". Changing "chains.txt" is an easy way to test different compositions and doesn't require regenerating the features.
In part based on: https://github.com/kalininalab/alphafold_non_docker
Installation will take around 1-2 hours. Tested on Linux (CentOS 7/8).
conda create --name alphalink -c conda-forge python=3.10
conda activate alphalink
For Linux:
pip install nvidia-pyindex
pip install https://github.com/dptech-corp/Uni-Core/releases/download/0.0.3/unicore-0.0.1+cu118torch2.0.0-cp310-cp310-linux_x86_64.whl
For other systems, build Uni-Core from scratch.
conda install -y -c conda-forge openmm==7.7.0 pdbfixer biopython==1.81
conda install -y -c bioconda hmmer hhsuite==3.3.0 kalign2
pip install tensorflow-cpu==2.16.1
git clone https://github.com/deepmind/alphafold.git
cd alphafold
python setup.py install
# download folding resources
wget --no-check-certificate https://git.scicore.unibas.ch/schwede/openstructure/-/raw/7102c63615b64735c4941278d92b554ec94415f8/modules/mol/alg/src/stereo_chemical_props.txt
# copy stereo_chemical_props.txt to the alphafold conda folder
cp stereo_chemical_props.txt $CONDA_PREFIX/lib/python3.10/site-packages/`ls $CONDA_PREFIX/lib/python3.10/site-packages/ | grep alphafold`/alphafold/common/
cd ..
If you are missing the databases for MSA generation, you can download them with the following script:
bash scripts/download/download_all_data.sh /path/to/database/directory full_dbs
or for the smaller databases:
bash scripts/download/download_all_data.sh /path/to/database/directory reduced_dbs
They require up to 3TB of storage.
git clone https://github.com/Rappsilber-Laboratory/AlphaLink2.git
cd AlphaLink2
python setup.py install
The model weights are deposited here:
After set up, AlphaLink can be run as follows:
bash run_alphalink.sh \
/path/to/the/input.fasta \ # target fasta file
/path/to/crosslinks.pkl.gz \ # pickled and gzipped dictionary with crosslinks
/path/to/the/output/directory/ \ # output directory
/path/to/model_parameters.pt \ # model parameters
/path/to/database/directory/ \ # directory of databases
2020-05-01 # use templates before this dateOutput folder will contain the relaxed and unrelaxed PDBs and a pickle file with the PAE map.
We expose also 4 optional parameters to set the number of recycling iterations, number of samples, Neff for subsampling MSAs, and the possibility to remove MSA information for crosslinked residues.
bash run_alphalink.sh \
/path/to/the/input.fasta \ # target fasta file
/path/to/crosslinks.pkl.gz \ # pickled and gzipped dictionary with crosslinks
/path/to/the/output/directory/ \ # output directory
/path/to/model_parameters.pt \ # model parameters
/path/to/database/directory/ \ # directory of databases
2020-05-01 \ # use templates before this date
20 \ # use 20 recycling iterations (default: 20)
25 \ # generate 25 sample (default: 25)
30 \ # downsample MSAs to Neff 30 (default: -1, use full MSA, expects integer >= 1)
1 # integer > 0 activates this option. Remove MSA information for crosslinked residues (default: -1, use full MSA)Models generated with AlphaLink using experimental restraints can be published as integrative/hybrid models in PDB-Dev PDB-Dev using the make_ihm.py script which requires python-ihm.
The script takes the chain_id_map.json file, the crosslink pickle, a mmcif file generated from the .pdb output of AlphaLink2 and the accession code for the deposited data (e.g., PRIDE) as input.
To generate a mmcif file from the .pdb output of AlphaLink2 you can use Maxit.
Finally update the authors in the make_ihm.py script and if applicable add your publication as a citation before running the script.
GPU, ideally NVIDIA V100 and upwards. A100+ can make use of bfloat16 to predict larger targets.
If you use the code, the model parameters, or the released data of AlphaLink2, please cite
@article{stahl2024modelling,
title={Modelling protein complexes with crosslinking mass spectrometry and deep learning},
author={Stahl, Kolja and Warneke, Robert and Demann, Lorenz and Bremenkamp, Rica and Hormes, Bj{\"o}rn and Brock, Oliver and St{\"u}lke, J{\"o}rg and Rappsilber, Juri},
journal={Nature communications},
volume={15},
number={1},
pages={7866},
year={2024},
publisher={Nature Publishing Group UK London}
}Any work that cites AlphaLink2 should also cite AlphaFold and Uni-Fold.
AlphaLink2 is based on Uni-Fold and fine-tunes the network weights of AlphaFold.
While AlphaFold's and, by extension, Uni-Fold's source code is licensed under the permissive Apache License, Version 2.0, DeepMind's pre-trained parameters fall under the CC BY 4.0 license. Note that the latter replaces the original, more restrictive CC BY-NC 4.0 license as of January 2022
The AlphaLink parameters are made available under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) license. You can find details at: https://creativecommons.org/licenses/by/4.0/legalcode
Use of the third-party software, libraries or code referred to in the Acknowledgements section above may be governed by separate terms and conditions or license provisions. Your use of the third-party software, libraries or code is subject to any such terms and you should check that you can comply with any applicable restrictions or terms and conditions before use.