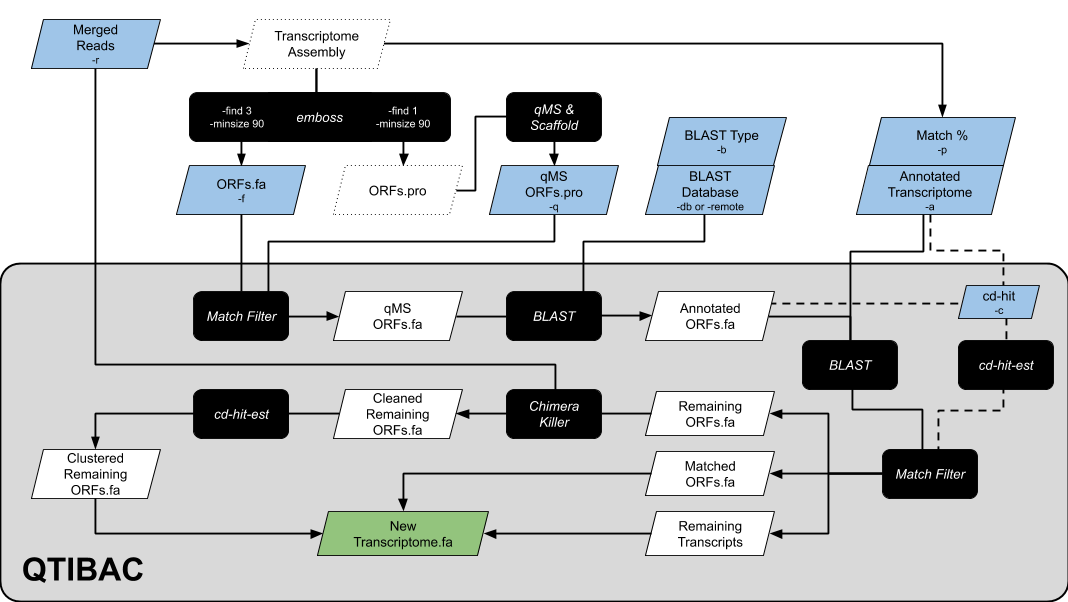
QMS-based Transcript Identification, Blast, Annotation, and Confirmation
QTIBAC is a wrapper script which uses blast, cd-hit, and ChimeraKiller to cluster proteomically (quantitative mass spectrometry/qMS) identified ORFs to your transcriptome, blast them to a provided database (or to the NR database on Genbank), and remove chimeric ORFs.
QTIBAC provides a new transcriptome with (1) proteomically confirmed transcripts in your transcriptome designated with *** and (2) non-matched proteomically identifed ORFs appended as 'Putative' novel transcripts.
- You have used
embossto rungetorf -find 1 -minsize 90 -sequence {}_assembly.fasta -outseq {}_ORFs.pro - You have used
embossto rungetorf -find 3 -minsize 90 -sequence {}_assembly.fasta -outseq {}_ORFs.fasta - You have used
{}_ORFs.proas your input for qMS identification and quantification.
conda create -n qtibac_env python=3.6 biopython blast cd-hit bwa samtools bedtools picard pandas matplotlib scipy pysam gatk4 pathos
git clone https://github.com/RhettRautsaw/QTIBAC.git
git clone https://github.com/masonaj157/ChimeraKiller.git
echo 'export PATH=$PATH:/path/to/QTIBAC:/path/to/ChimeraKiller' >> ~/.bash_profile
-f {fasta}: nucleotide ORFs made usingemboss(see Assumptions section; (e.g.,-f {}_ORFs.fasta)-q {fasta}: qMS matched protein ORFs, exported fromScaffold4or a similar program (e.g.,-q {}_Scaffold.pro)-a {fasta}: annotated transcriptome fasta (e.g.,-a {}_transcriptome_v1.fasta)-r {fastq}: PEAR merged reads fastq (e.g.,-r {}_pear.fastq)- Choose option below
-db {path}&-b {blastn/blastx}: User specified blast database (withmakeblastdbpreviously run) and the type ofblastsearch (e.g.,-db /path/to/swissprot -b blastx)-remote: blast against the GenBank nr database instead (Warning: Much slower)
-o {string}: name for output files-c: usecd-hitinstead ofblastwhen clustering to your transcriptome-p {integer}:blastorcd-hitmatch percentage when comparing ORFs to transcriptome (default = 99%)-t {integer}: number of processing threads (default = 8)
-h, --help show this help message and exit
-f FASTA, --fasta FASTA
REQUIRED: Nucleotide ORFs fasta made from assembly and matching the protein ORFs used for QMS identification (i.e. emboss::getorf -find 3)
-q QMS, --qms QMS
REQUIRED: qMS protein fasta (exported from Scaffold4)
-a ANNOTATED, --annotated ANNOTATED
REQUIRED: Annotated transcriptome fasta
-r READS, --reads READS
REQUIRED: PEAR merged reads
-db BLAST_DATABASE, --blast_database BLAST_DATABASE
Path to blast database for annotation (e.g. SWISSprot). Assumes makeblastdb has already been run.
-b BLAST_TYPE, --blast_type BLAST_TYPE
Type of blast to run (e.g. blastn or blastx) against blast database for annotation. Default assumes blastx
-remote Use remote blast against nucleotide database instead of a local blast database. WARNING: This will take much much much much much longer!
-c, --cdhit Use cd-hit-est instead of blastn to compare to your annotated transcriptome
-o OUTPUT, --output OUTPUT
Conserved part of name for output files
-p [MATCHPERCENT], --matchpercent [MATCHPERCENT]
Integer percent identity for blastn or cd-hit-est. Default is 99
-t NUM_THREADS, --num_threads NUM_THREADS
Number of threads for blast. Default is 8
-bp BLAST_PATH, --blast_path BLAST_PATH
Directory with blast command. Default assumes it is in your path (e.g. /PATH/TO/BIN/WITH/BLAST/)
-mbdb MAKEBLASTDB, --makeblastdb MAKEBLASTDB
Directory with makeblastdb command. Default assumes it is in your path (e.g. /PATH/TO/BIN/WITH/MAKEBLASTDB/)
-cdp CDHIT_PATH, --cdhit_path CDHIT_PATH
Directory with cd-hit-est-2d command. Default assumes it is in your path (e.g. /PATH/TO/BIN/WITH/CDHIT/)
--version show program's version number and exit
- Fasta of nucleotide ORFs matching QMS proteins
{}_QMS_DB.fasta - Fasta of nucleotide ORFs with first BLAST hit in description
{}_QMS_hits.fasta - An xml of top 10 BLAST hits
{}_blast.xml - Fasta of QMS transcripts which matched to the annotated transcriptome
- Fasta of QMS transcripts which remain after failing to match the annotated transcriptome
- Fasta of QMS transcripts which remain after removing chimeric sequences
- Fasta of QMS transcripts which remain after clustering clean sequences
- New annotated transcriptome with *** indicating which transcripts are proteomically confirmed and remaining orfs appended
Blasting to local database and comparing to annotated transcriptome using cd-hit (note the -db -b -c flags)
QTIBAC.py -f Cline-CLP2629_ORFs.fasta -q Cline-CLP2629_Scaffold.pro -a Cline-CLP2629_transcriptome_v3.fasta -r Cline-CLP2629_M.assembled.fastq.gz -db /PATH/TO/SWISSprot -b blastx -c -o Cline-CLP2629 -p 99 -t 8
Blasting to remote database and comparing to annotated transcriptome using blast (note the -remote flag and lack of -c flag)
QTIBAC.py -f Cline-CLP2629_combined_ORFs.fasta -q Cline-CLP2629_QMS_DB.pro -a Cline-CLP2629_transcriptome_v3.fasta -r Cline-CLP2629_M.assembled.fastq.gz -remote -o Cline-CLP2629 -p 99 -t 8