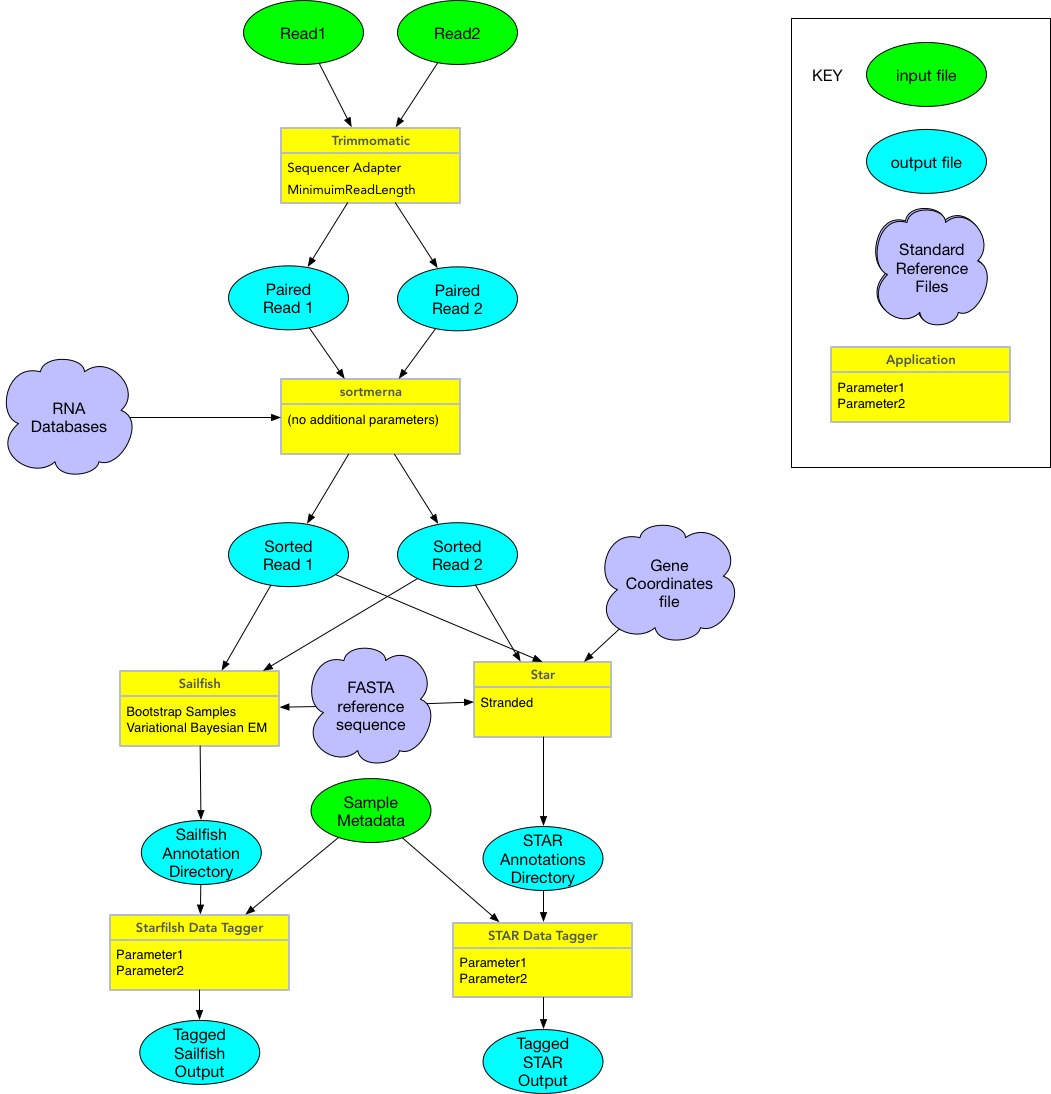
#RNASeq reactor
This ETL step runs up to two applications on single or double read sequences:
- Trimmomatic to trim raw reads
- Sortmerna to remove known sequences
There three inputs needed and four options in the json file. Inputs are
- read1 the fastq of the first (or only) sequence read
- read2 the fastq of the second read (or to signal there is no read2 use the empty file agave://data-sd2e-community/sample/rnaseq/test/ENOSUCH_R2.fastq.gz)
- filterseqs The directory with the databases of rna sequences to reject in the format used by sortmerna in a single directory plus a file named config which has the command argument to pass to sortmerna
The default filterseqs from Q0 is located at agave://data-sd2e-community/reference/rna_databases/rRNA_databases
Note the filterseqs should have no path information to the files but should have all the other information in the format of
rRNA_databases/silva-bac-16s-id90.fasta,rRNA_databases/silva-bac-16s-db:
rRNA_databases/silva-bac-23s-id98.fasta,rRNA_databases/silva-bac-23s-db:
rRNA_databases/silva-arc-16s-id95.fasta,rRNA_databases/silva-arc-16s-db:
rRNA_databases/silva-arc-23s-id98.fasta,rRNA_databases/silva-arc-23s-db:
rRNA_databases/silva-euk-18s-id95.fasta,rRNA_databases/silva-euk-18s-db:
rRNA_databases/silva-euk-28s-id98.fasta,rRNA_databases/silva-euk-28s:
rRNA_databases/rfam-5s-database-id98.fasta,rRNA_databases/rfam-5s-db:
rRNA_databases/rfam-5.8s-database-id98.fasta,rRNA_databases/rfam-5.8s-db
The options for this applicaiton are
- trim (true,false) run trimmomatic on the read(s)
- adaptersfile the adapter file used for trimming (Q0 value was TruSeq3-PE.fa:2:30:10)
- minlen the minlen value passed to sortmerna
- sortmerna (true,false) run the sortmerna step
Note all three inputs and four options are required even if no processing is to be done (this will be addressed in a future release of agave). Also all inputs must exist and be readable within the SD2E environment.
When run, all cores available to the VM will be used for the processing. The applicaition will autodetect the number of cores.
The outputs from the two steps of this proces (trimmomatic and sortmerna) are R1 and R2 files for each input pair plus errors and log files
- _paired.fastq.gz: for each paired end, from trimmomatic
- _rna_free_reads.fastq: for each paired end, from sortmeRNA
- <job_name>-<job_id>.err: trimmomatic: reports # and % paired reads, # and % dropped
- <job_name>-<job_id>.log: the log file for the overall job
Examples of the job script can be found in the rnaseq-0.1.2 directory *rnaseq-job.json.
Once completed, additional post processing steps are carried out to annotate the sequences. These are separate applications, starfish and rnaseq-broad, which can be run on any pairs of sorted reads.