Python package to process EEG data from BrainVision's 'Analyzer' and 'Recorder'.
Still in development.
I will develop testings on next update.
python setup.py install --record files.txt
cat files.txt | xargs rm -rf
- numpy
- scypy
from data_structures import EEGData
eeg_data = EEGData('./SampleData/sample.mat')
print(eeg_data.channel_names)
print(eeg_data.properties)
print(eeg_data.markers)
print(eeg_data.signals)To save using memory, I revised interfaces using intermediate data.
Please look below and tests/* or ~example.
Calculate Phase locking factor and nonparametric testing.
Referenced
Oscillatory gamma-band (30-70 Hz) activity induced by a visual search task in humans (Tallon et al. 1997)
Please see the document if you want more details.
There is an information about 'sig_name' in Appendix.
'trial_marker' is a marker you decided to represent the start of the trials.
show_plf_spectgram returns plf matrix and p values matrix which are type of numpy.array.
The p values are calculated to test whether an activity is significantly phase-locked to stimulus onset,
a statistical test (Rayleigh test) of uniformity of angle is used (Jervis et al., 1983)
Firstly, you need to transform original data to wavelet transformed signals.
import numpy as np
import scipy.io as sio
from pyplfv.data_structures import EEGData
from pyplfv.wavelet import save_waveleted_signal
from pyplfv.wavelet import save_waveleted_signal_with_farray
from pyplfv.wavelet import save_waveleted_eegdata_with_farray
'''
def save_waveleted_signal(signal, sampling_interval, f0, filename):
def save_waveleted_signal_with_farray(signal, sampling_interval, farray, path):
def save_waveleted_eegdata_with_farray(eegdata, sampling_interval, farray, path):
'''
matdata = sio.loadmat('SampleData/simulation_data1.mat')
trial_num = len(matdata['seg'][0])
arr = np.array(matdata['seg'], dtype='float128')
arr = arr.T
sig = np.hstack([arr[i] for i in range(trial_num)])
time_interval = 0.002
farray = [1.0 * i for i in range(1,30)]
start_time_of_trials = [750 * i for i in range(trial_num)]
offset = 0
length = int(1.5 / 0.002)
save_waveleted_signal_with_farray(signal=sig, sampling_interval=time_interval, farray=farray, path='./SampleData/simulationData1')Next, transform wavelet signals to normalized TVE or TVE.
from pyplfv.tve import save_normalized_tve_with_farray
from pyplfv.tve import save_tve_with_farray
from pyplfv.tve import save_tve_of_eegdata_with_farray
save_tve_with_farray(waveleted_path='SampleData/simulationData1')
save_normalized_tve_with_farray(waveleted_path='SampleData/simulationData1')
save_tve_of_eegdata_with_farray(waveleted_eegdata_path='SomeWaveletTransformedEEGDatapath')Finaly, transform normalized tve to PLF.
import numpy as np
from pyplfv.plf import save_plf_with_farray
#Save
trial_num = 100
farray = [1.0 * i for i in range(10,30)]
start_time_of_trials = [750 * i for i in range(trial_num)]
offset = 0
length = int(1.5 / 0.002)
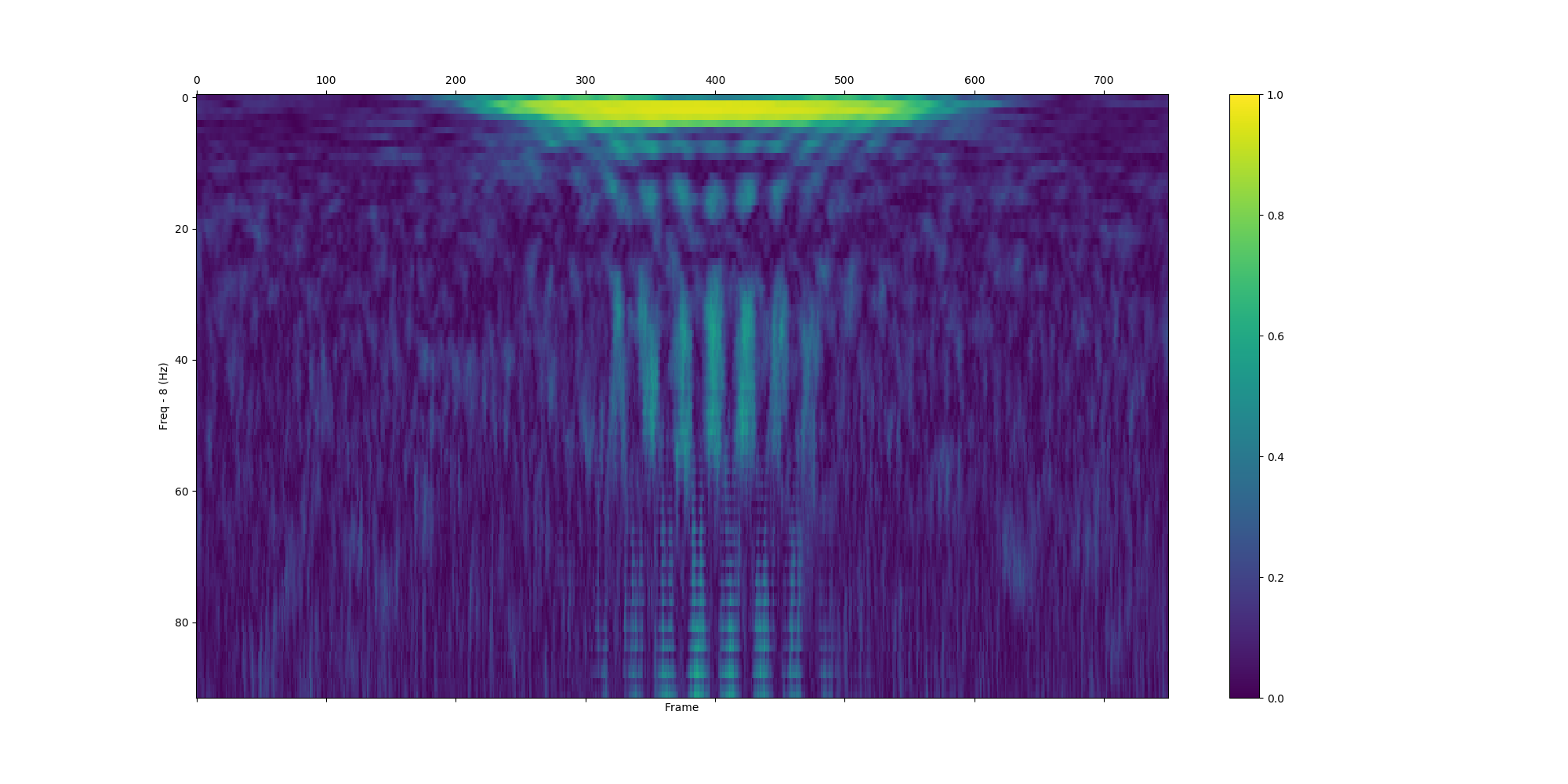
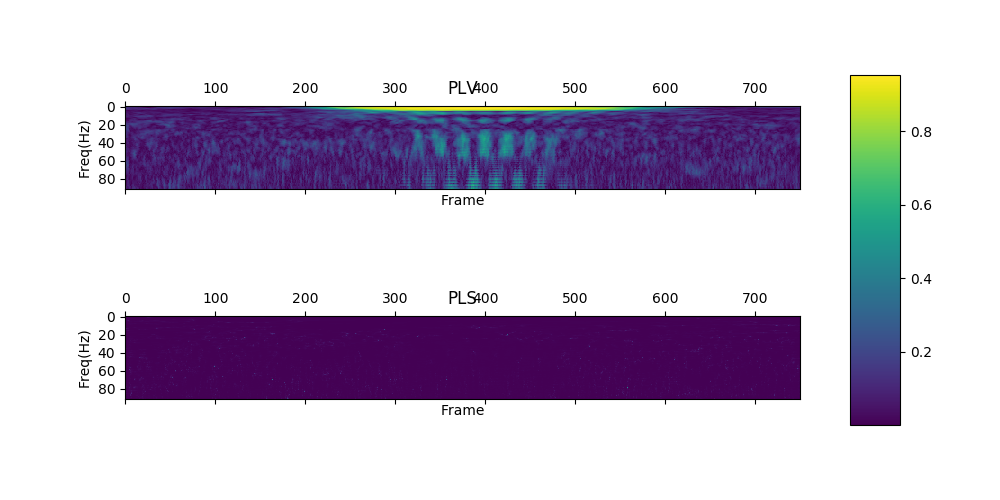
save_plf_with_farray('SampleData/simulationData1', start_time_of_trials, offset, length)| Pseudo random added 10Hz gaussian | Pseudo random added 50Hz gaussian | |
|---|---|---|
| signals |  |
 |
| PLF | 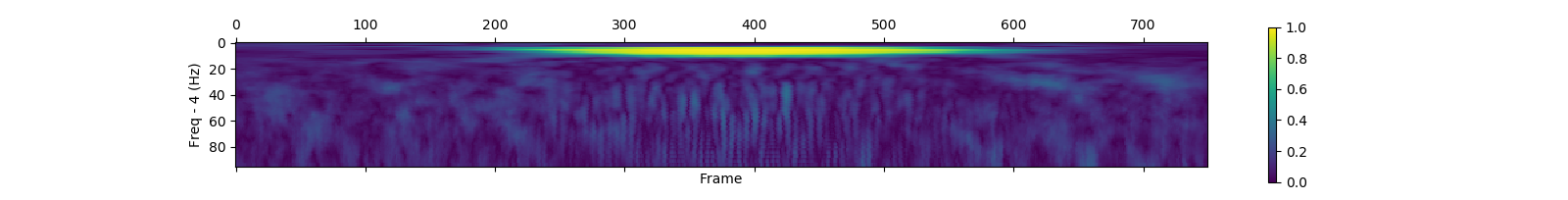 |
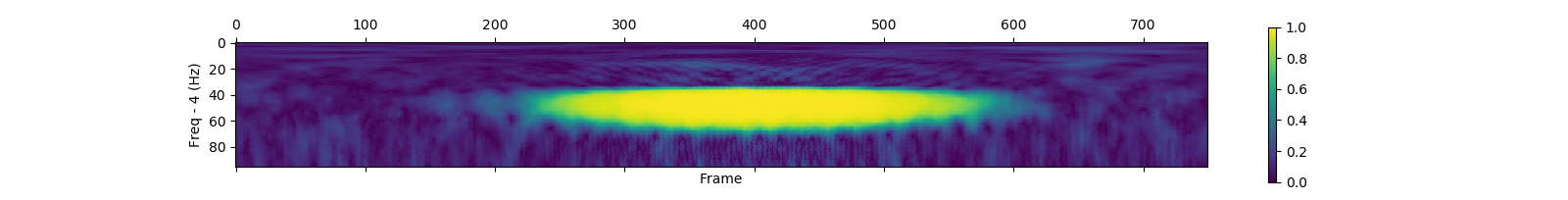 |
| PLF /w p values | 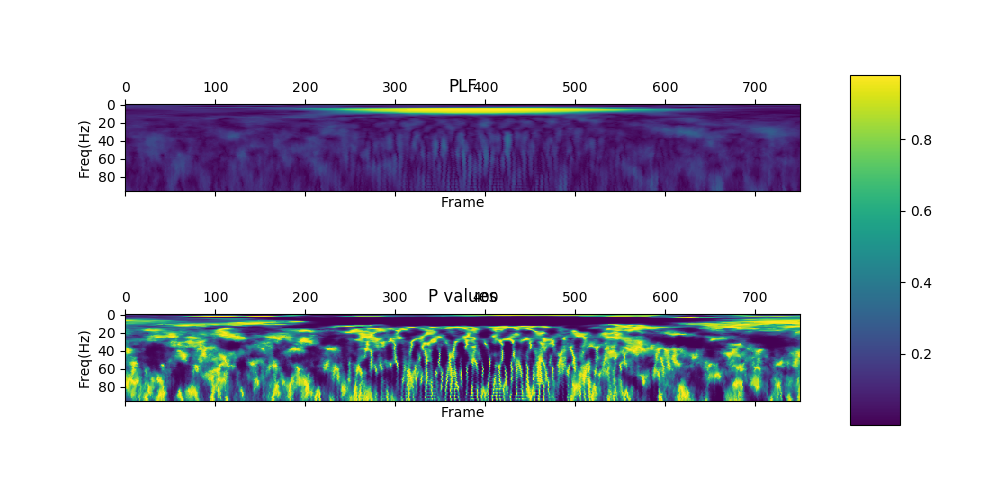 |
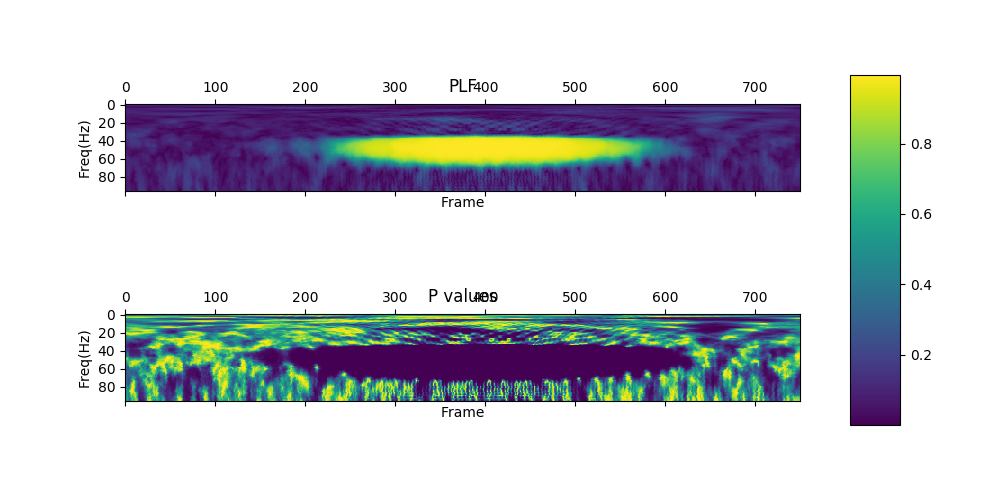 |
If you want more samples, please see 'plf_samples.py'.
Calculate Phase locking value and statistical test based on surrogate data.
Referenced
Measuring Phase Synchrony in Brain Signals (Lachaux et al. 1997)
Please see the document if you want more details.
plv_bet_2ch returns plv matrix which is type of numpy.array.
When you want to get the plv about each situations, you can add parameters 'trial_filering' and 'trial_filter'.
If only you need is plv matrix, you can hide the heatmap by make show_mat=False.
Firstly, you need to transform original data to wavelet transformed signals like as above.
And next, you choose interface corresponding your data structures.
from data_structures import EEGData
from plv import plv_bet_2ch
eeg_data = EEGData('./SampleData/sample.mat')
#def plv_bet_2ch(eeg_data, sig_name1, sig_name2, trial_marker, farray, offset, length, trial_filering=False, trial_filter=[], show_mat=False):
plvs = plv_bet_2ch(eeg_data, 'Pz', 'Cz', 'S255', np.arange(15.0, 80.0, 1.0), int(-1.0 / 0.002), int(3 / 0.002))or
situation_0 = [1, 4, 8, 13, 15, 21, 23, 25, 26, 30, 31, 32, 38, 40, 44, 47, 52, 59, 62, 64, 65, 82, 85, 89, 92, 99, 105, 107, 112, 114, 117, 133, 138, 144, 145, 146, 147, 149]
situation_7 = [0, 9, 11, 12, 14, 27, 29, 33, 36, 39, 43, 46, 53, 56, 60, 61, 63, 71, 74, 75, 81, 90, 93, 96, 104, 106, 108, 109, 113, 116, 118, 120, 124, 126, 127, 129, 132, 135, 136, 140, 142, 148]
plv_bet_2ch(eeg_data, 'Pz', 'Cz', 'S255', np.arange(15.0, 80.0, 1.0), int(-1.0 / 0.002), int(3 / 0.002), True, situation_0, True)
plv_bet_2ch(eeg_data, 'Pz', 'Cz', 'S255', np.arange(15.0, 80.0, 1.0), int(-1.0 / 0.002), int(3 / 0.002), True, situation_7, True)| All trials | Situation_0 | Situation_7 |
|---|---|---|
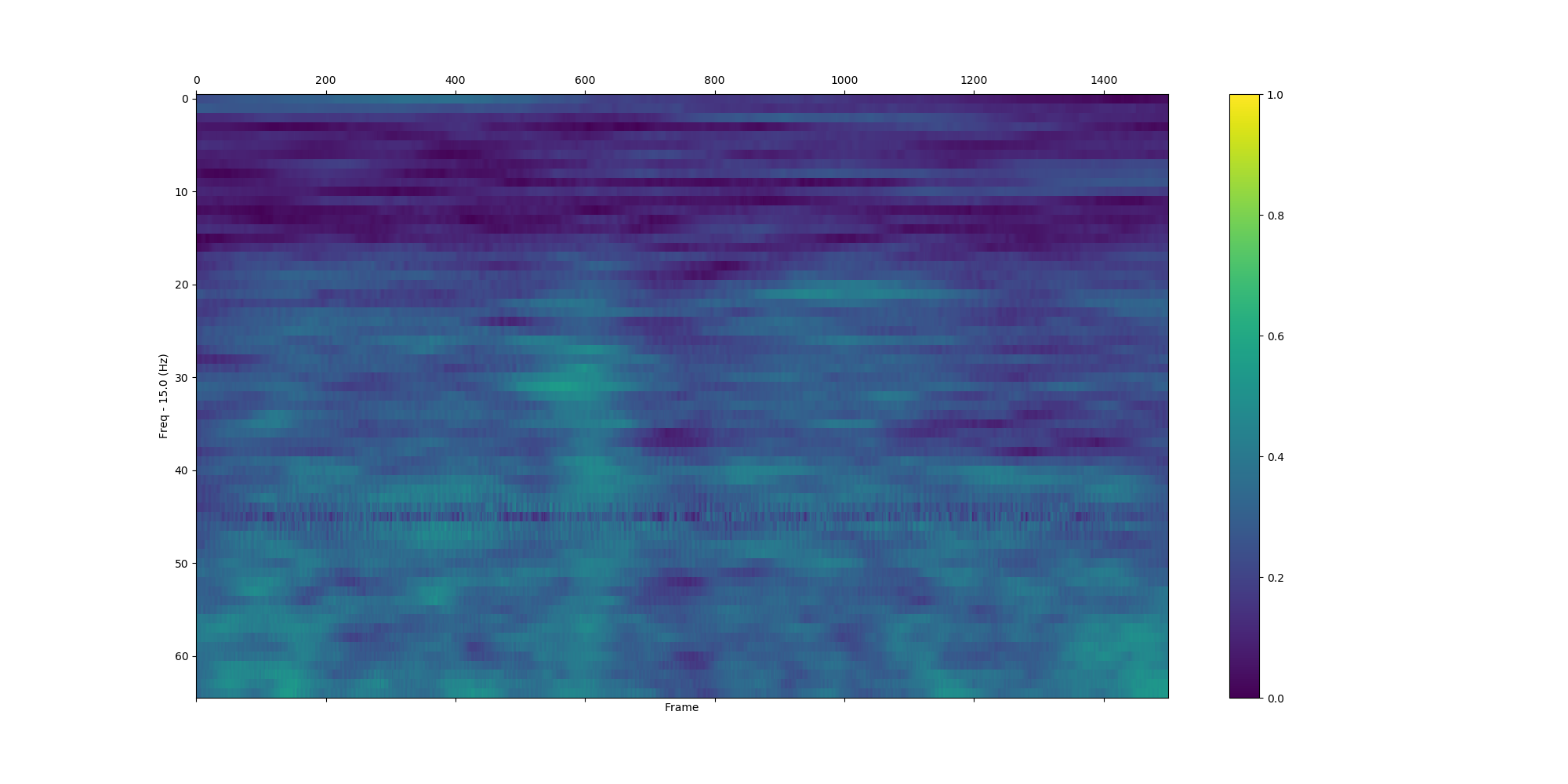 |
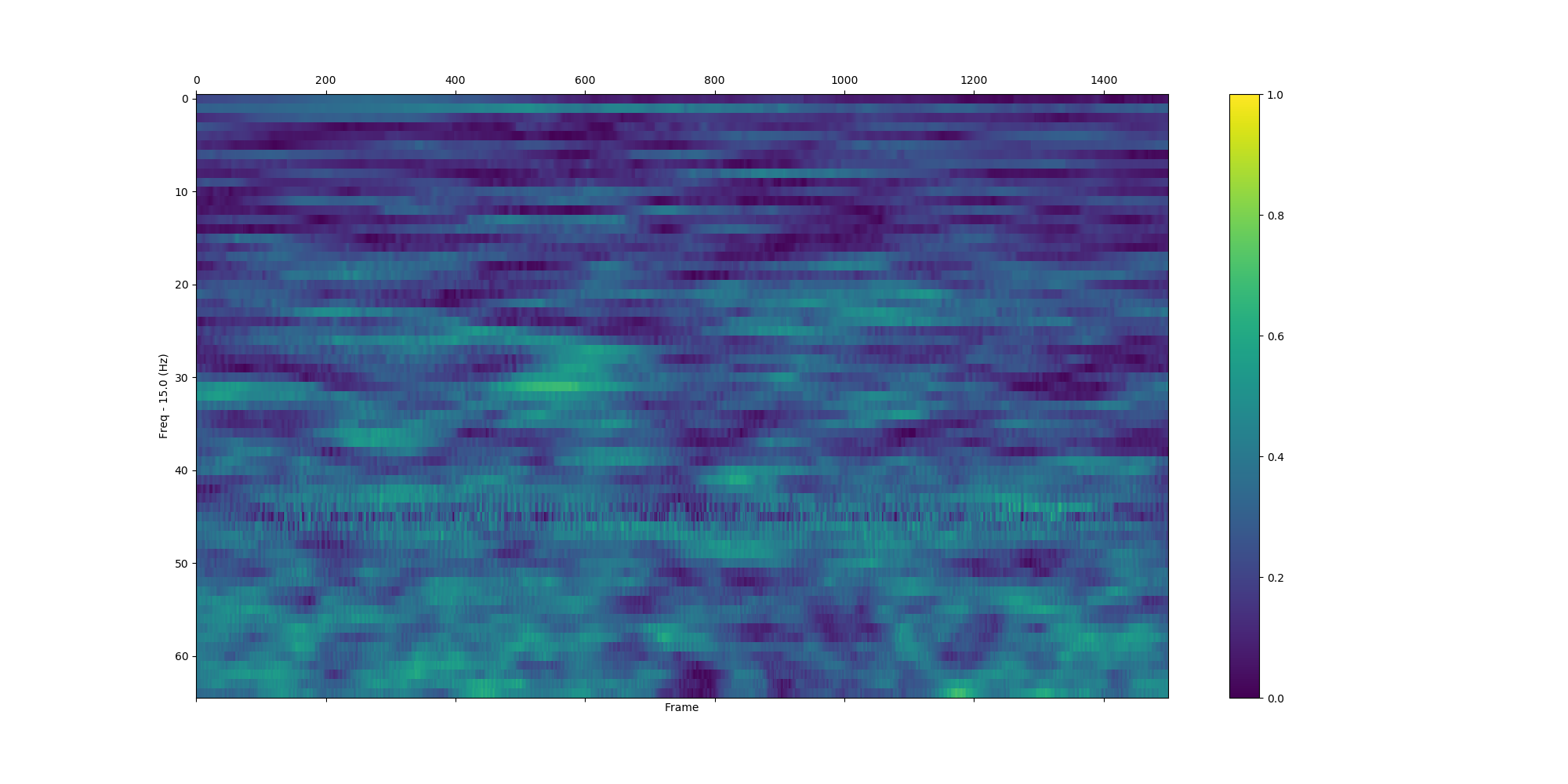 |
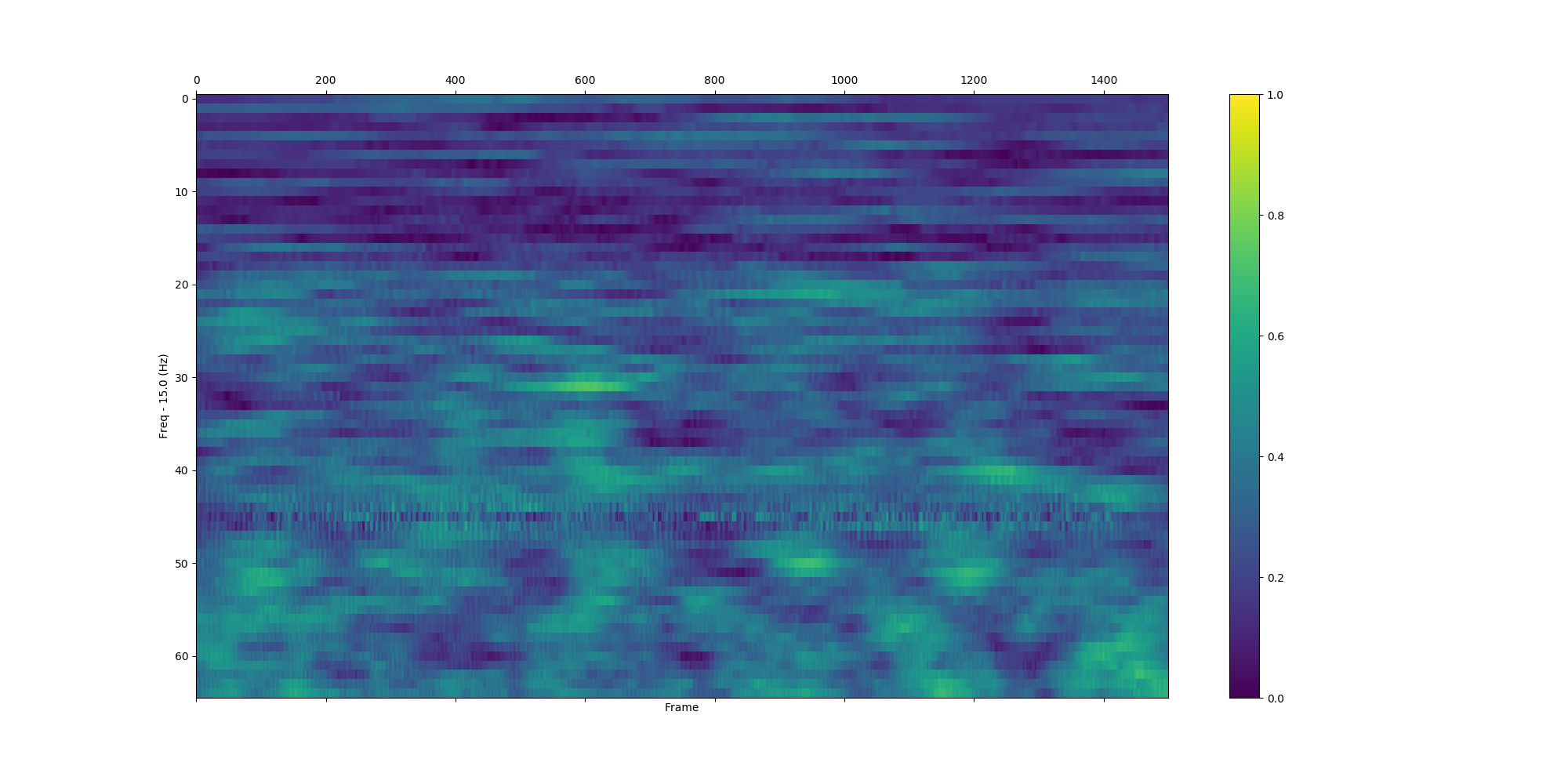 |
or
# plv_bet_2ch(ch1, ch2, time_interval, start_time_of_trials, farray, offset, length, show_test=False, save=False, filename='Images/plv.png'):
matdata = sio.loadmat('SampleData/simulation_data1.mat')
trial_num = len(matdata['seg'][0])
arr = np.array(matdata['seg'], dtype='float128')
arr = arr.T
ch1 = np.hstack([arr[i] for i in range(trial_num)])
matdata = sio.loadmat('SampleData/simulation_data2.mat')
trial_num = len(matdata['seg'][0])
arr = np.array(matdata['seg'], dtype='float128')
arr = arr.T
ch2 = np.hstack([arr[i] for i in range(trial_num)])
farray = [1.0 * i for i in range(4,100)]
start_time_of_trials = [750 * i for i in range(trial_num)]
offset = 0
length = int(1.5 / 0.002)
plvs, plss = show_plv_bet_2ch(ch1, ch2, 0.002, start_time_of_trials, farray, offset, length, True, True, 'Images/plv_simulation_wtest.png')Simulation_data1 and simulation_data2 which are inserted 10Hz.
These signals are perfectly phase-locked.
With surrogate data test.
MatData has these columns. Details are written below.
- 'Analyzer' : Analyze information on BrainVision's 'Analyzer'
- 'EEGData' : RealData
- 'EEGPoints' : Number of points
- 'EEGPosition' : ?
- 'FileName' : filename
- 'Markers' : markers which are inserted offline and online
- 'NodeName' : Node name in BrainVision's 'Analyzer'
- 'Properties' : Recording properties
'Fp1', 'Fp2', 'F3', 'F4', 'C3', 'C4', 'P3', 'P4', 'O1', 'O2', 'F7', 'F8', 'T7', 'T8', 'P7', 'P8', 'Fz', 'Cz', 'Pz', 'FC1', 'FC2', 'CP1', 'CP2', 'FC5', 'FC6', 'CP5', 'CP6', 'FCz', 'HEOGL', 'HEOGR', 'VEOGU', 'VEOGL'
Marker which are inserted offline and online.
In my case, output to recorder 'S255' to represent starts of trials.
And 'Bad Min-Max' markers are inserted by Analyzer.