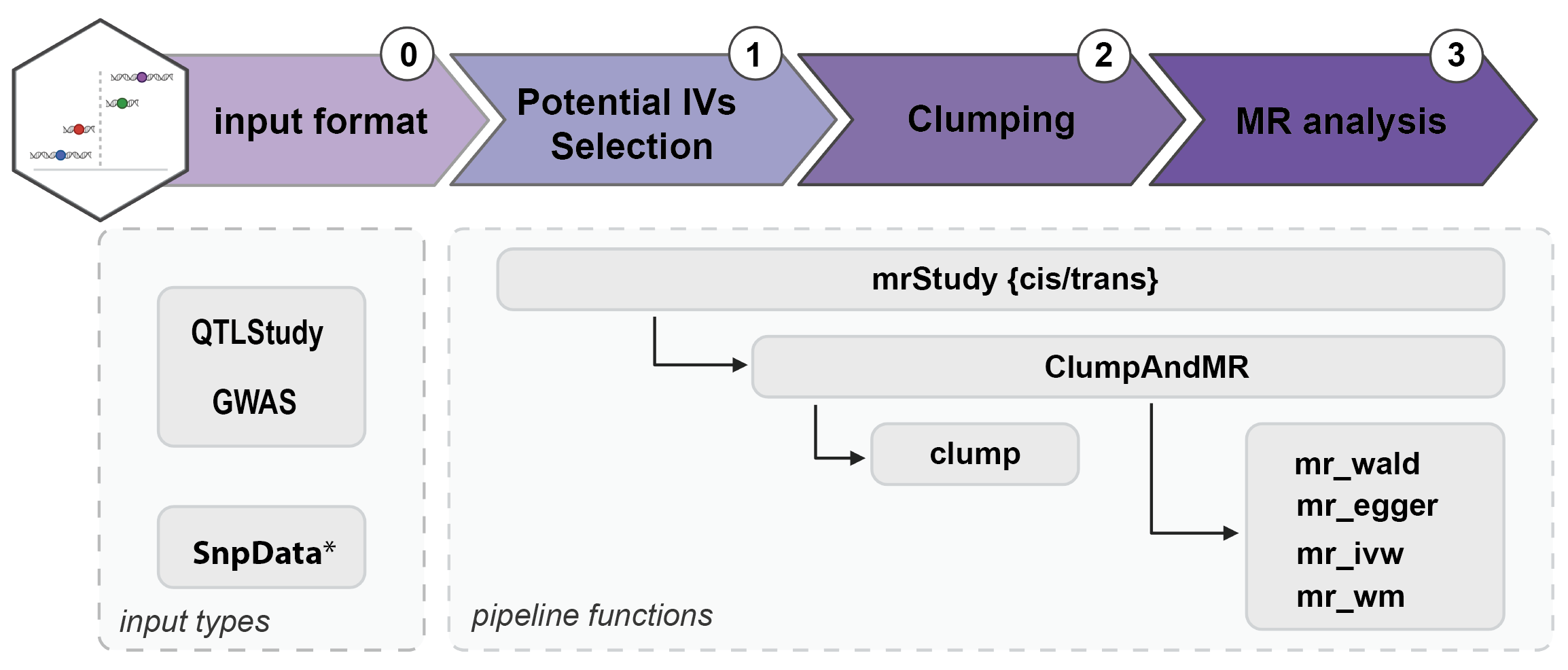
LaScaMolMR.jl (Large Scale Molecular Mendelian Randomization) is a threaded Mendelian Randomization (MR) package that is focused on the generation of transcriptome wide / molecular MR analysies. Although it provides interface for most common MR regression estimators (Inverse Variance Weighted, Weighted Median, Egger, Wald), its intended use is to enable fast Omic-wide Mendelian Randomization studies. The rise of large genetic cohort data has benefited the statistical power of Genome Wide Association Studies (GWAS) and Quantitative Trait Loci (QTL). Thus enabling findings in extensive studies such as Transcriptome Wide MR (TWMR), or mediation analyses between different levels of phenotypes. LaScaMolMR.jl provides a fast and efficient framework (still under developpement) to such analyses, allowing users to choose parameters of the study.
*SnpData is implemented in SnpArrays Package.
julia> ]
(@v1.10) pkg> add "https://github.com/SamuelMathieu-code/LaScaMolMR.jl"
For a QTL dataset composed as follows with a single file :
base/folder
└── all_explosures.txt
With example data composed like this (tab separated) :
chr pos A1 A2 beta se some_useless_column pval
1 10511 A G -0.176656 0.136297 . 0.194939
1 10642 A G -0.724554 0.345390 .. 0.035924
1 11008 G C -0.017786 0.016673 ... 0.286088
1 11012 G C -0.017786 0.016673 .. 0.286088
1 13110 A G 0.013272 0.021949 . 0.545411
1 13116 G T -0.027802 0.013111 .. 0.0339672
1 13118 G A -0.027802 0.013111 ... 0.0339672
1 13259 A G -0.122207 0.210776 .. 0.562052
1 13273 C G 0.007077 0.015337 . 0.644463
and a GWAS of outcome composed similarly but comma separated, the following code will generate a trans-MR study with default parameters :
using LaScaMolMR- Describe exposure data.
path_pattern = ["all_exposures.txt"]
columns = Dict(1 => CHR, 2 => POS, 3 => A_EFFECT, 4 => A_OTHER, 5 => BETA, 6 => SE, 8 => PVAL)
trait_v = ["A", "B", "C"] # Chromosome and TSS informations are not relevant in Trans setting.
exposure::QTLStudy = QTLStudy_from_pattern("base/folder/",
path_pattern,
trait_v, chr_v = nothing,
tss_v = nothing,
columns, separator = '\t',
only_corresp_chr = false)Of note, the path_pattern variable can adapt to other file achitectures, when exposures are dispacted in different files (see the full documentation).
- Describe outcome data.
outcome = GWAS("/some/file", columns, separator = ',', trait_name = "Some Painful Disease")- Perform Medelian Randomization study by providing input formats and reference genotype data files.
# Plink 1.9 files base names for each chromosome (You can also use a single file)
plink_files = ["folder/basename_file_chr$(i)" for i in 1:22]
# Perform MR for every exposure - outcme pairs with default parameters
out_table = mrStudy(exposure, outcome, "cis", plink_files)
# with MiLoP approach and other parameters :
out_table2 = mrStudy(exposure, outcome, "cis", plink_files,
approach = "MiLoP",
r2_tresh = 0.01,
p_tresh = 5e-8,
filter_beta_ratio = 1)
# Default p_tresh_MiLoP value is the same as p_treshMitigated Local Pleiotropy (MiLoP) approach modifies the potential IV selection process to remove instrument variables associated to more than 1 exposure at p_tresh_MiLoP significance level.
- Multivariate MR & TWMR according to Porcu et al.
- Steiger for causal direction assesment Hmani et al.
- Mediation analysis inspired by Auwerx et al.
- Web interface for locus visualization with Genie.jl