Official implementation of Generative Flows on Synthetic Pathway for Drug Design by Seonghwan Seo, Minsu Kim, Tony Shen, Martin Ester, Jinkyu Park, Sungsoo Ahn, and Woo Youn Kim. [arXiv]
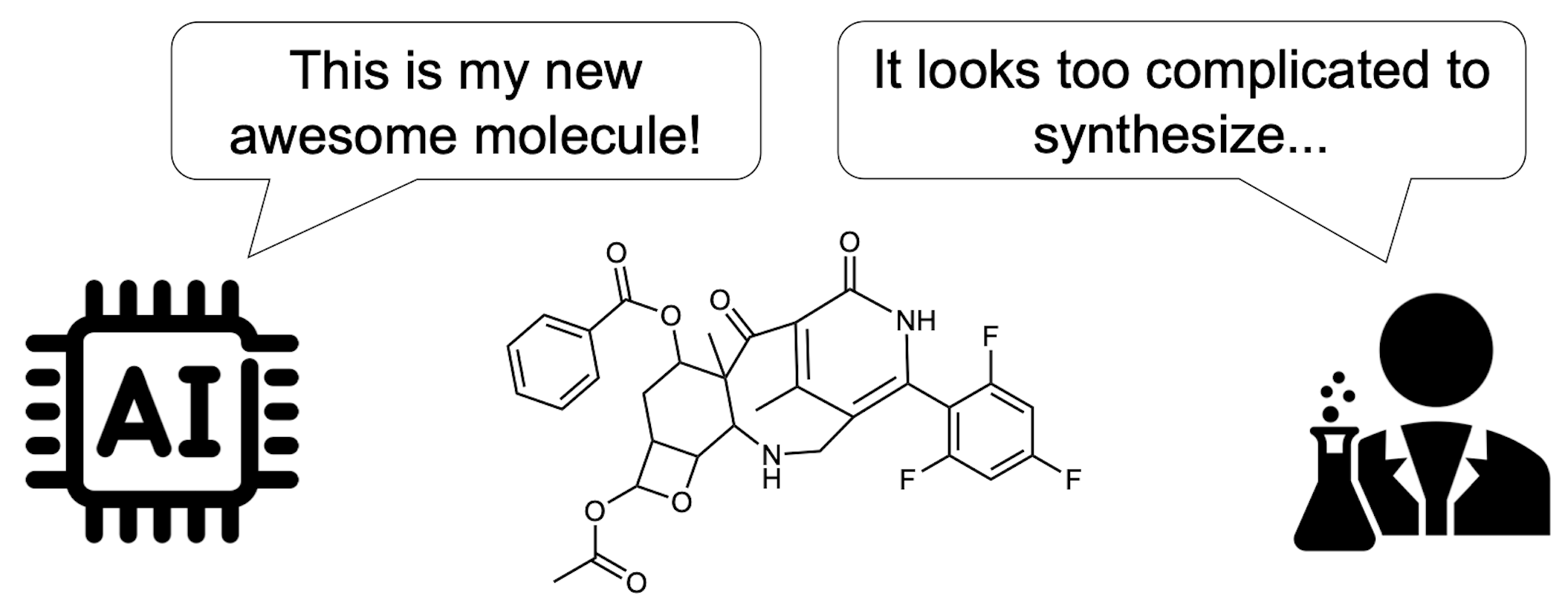
RxnFlow are a synthesis-oriented generative framework that aims to discover diverse drug candidates through GFlowNet objective and a large action space.
- RxnFlow can operate on large synthetic action spaces comprising 1.2M building blocks and 71 reaction templates without compute overhead
- RxnFlow can explore broader chemical space within less reaction steps, resulting in higher diversity, higher potency, and lower synthetic complexity of generated molecules.
- RxnFlow can generate molecules with expanded or modified building block libaries without retraining.
This project is based on gflownet, and src/gflownet/ is a clone of recursionpharma/gflownet@v0.2.0. Since we have updated the gflownet version and performed modularization after submission, we do not guarantee that current version will reproduce the same results as the paper. You can access the reproducing codes and scripts from tag: paper-archive.
This repository was developed for research. The code for real-world drug discovery will be released later.
# python: 3.10
conda install openbabel # For PharmacoNet
pip install -e . --find-links https://data.pyg.org/whl/torch-2.3.1+cu121.html
# For UniDock
conda install openbabel unidock
pip install -e '.[unidock]' --find-links https://data.pyg.org/whl/torch-2.3.1+cu121.htmlTo construct the synthetic action space, RxnFlow requires the reaction template set and the building block library. We provide two reaction template set:
- The reaction template used in this paper contains 13 uni-molecular reactions and 58 bi-molecular reactions, which is constructed by Cretu et al. The template set is available under data/templates/hb_edited.txt.
- We provide the new reaction template set data/templates/real.txt from Enamine REAL synthesis protocol.
The Enamine building block library is available upon request at https://enamine.net/building-blocks/building-blocks-catalog. We used the "Comprehensive Catalog" released at 2024.06.10.
-
Use Comprehensive Catalog
cd data # case1: single-step python scripts/a_sdf_to_env.py -b <CATALOG_SDF> -d envs/enamine_all --cpu <CPU> # case2: two-step python scripts/b1_sdf_to_smi.py -b <CATALOG_SDF> -o building_blocks/blocks.smi --cpu <CPU> python scripts/b2_smi_to_env.py -b building_blocks/blocks.smi -d envs/enamine_all --cpu <CPU> --skip_sanitize python scripts/b2_smi_to_env.py -b building_blocks/blocks.smi -d envs/real -t tesmplates/real.txt --cpu <CPU> --skip_sanitize
-
Use custom SMILES file (
.smi)python scripts/b2_smi_to_env.py -b <SMILES-FILE> -d ./envs/<ENV> -t ./templates/<RXN> --cpu <CPU>
You can optimize the docking score with GPU-accelerated UniDock.
python script/opt_unidock.py -h
python script/opt_unidock.py \
-p <Protein PDB path> \
-c <Center X> <Center Y> <Center Z> \
-l <Reference ligand, required if center is empty. > \
-s <Size X> <Size Y> <Size Z> \
-o <Output directory> \
-n <Num Oracles (default: 1000)> \
--filter <drugfilter; choice=(lipinski, veber, null); default: lipinski> \
--batch_size <Num generations per oracle; default: 64> \
--env_dir <Environment directory> \
--subsample_ratio <Subsample ratio; memory-variance trade-off; default: 0.01>You can also perform multi-objective optimization (Multi-objective GFlowNet) for docking score and QED.
python script/opt_unidock_moo.py -h
python script/opt_unidock_moo.py \
-p <Protein PDB path> \
-c <Center X> <Center Y> <Center Z> \
-l <Reference ligand, required if center is empty. > \
-s <Size X> <Size Y> <Size Z> \
-o <Output directory> \
-n <Num Oracles (default: 1000)> \
--batch_size <Num generations per oracle; default: 64> \
--env_dir <Environment directory> \
--subsample_ratio <Subsample ratio; memory-variance trade-off; default: 0.01>Example (KRAS G12C mutation)
-
Use center coordinates
python script/opt_unidock.py -p ./data/examples/6oim_protein.pdb -c 1.872 -8.260 -1.361 -o ./log/kras --filter veber python script/opt_unidock_moo.py -p ./data/examples/6oim_protein.pdb -c 1.872 -8.260 -1.361 -o ./log/kras --filter veber
-
Use center of the reference ligand
python script/opt_unidock.py -p ./data/examples/6oim_protein.pdb -l ./data/examples/6oim_ligand.pdb -o ./log/kras python script/opt_unidock_moo.py -p ./data/examples/6oim_protein.pdb -l ./data/examples/6oim_ligand.pdb -o ./log/kras
Not Implemented yet
Sample high-affinity molecules. The QuickVina docking score is estimated by Proxy Model [github].
python script/sampling_zeroshot.py -h
python script/sampling_zeroshot.py \
-p <Protein PDB path> \
-c <Center X> <Center Y> <Center Z> \
-l <Reference ligand, required if center is empty. > \
-o <Output path: `smi|csv`> \
-n <Num samples (default: 100)> \
--env_dir <Environment directory> \
--model_path <Checkpoint path; default: None (auto-downloaded)> \
--subsample_ratio <Subsample ratio; memory-variance trade-off; default: 0.01> \
--cudaExample (KRAS G12C mutation)
-
csvformat: save molecules with their rewards (GPU is recommended)python script/sampling_zeroshot.py -o out.csv -p ./data/examples/6oim_protein.pdb -l ./data/examples/6oim_ligand.pdb --cuda
-
smiformat: save molecules only (CPU: 0.06s/mol, GPU: 0.04s/mol)python script/sampling_zeroshot.py -o out.smi -p ./data/examples/6oim_protein.pdb -c 1.872 -8.260 -1.361
If you want to train RxnFlow with your custom reward function, you can use the base classes from rxnflow.base. The reward should be Non-negative.
-
Example (QED)
import torch from rdkit.Chem import Mol as RDMol, QED from gflownet import ObjectProperties from rxnflow.base import RxnFlowTrainer, RxnFlowSampler, BaseTask class QEDTask(BaseTask): def compute_obj_properties(self, objs: list[RDMol]) -> tuple[ObjectProperties, torch.Tensor]: fr = torch.tensor([QED.qed(mol) for mol in mols], dtype=torch.float).reshape(-1, 1) is_valid_t = torch.ones((len(mols),), dtype=torch.bool) return ObjectProperties(fr), is_valid_t class QEDTrainer(RxnFlowTrainer): # For online training def setup_task(self): self.task: QEDTask = QEDTask(cfg=self.cfg, wrap_model=self._wrap_for_mp) class QEDSampler(RxnFlowSampler): # Sampling with pre-trained GFlowNet def setup_task(self): self.task: QEDTask = QEDTask(cfg=self.cfg, wrap_model=self._wrap_for_mp)
Current version do not provide the reproducing code. Please switch to tag: paper-archive.
If you use our code in your research, we kindly ask that you consider citing our work in papers:
@article{seo2024generative,
title={Generative Flows on Synthetic Pathway for Drug Design},
author={Seo, Seonghwan and Kim, Minsu and Shen, Tony and Ester, Martin and Park, Jinkyoo and Ahn, Sungsoo and Kim, Woo Youn},
journal={arXiv preprint arXiv:2410.04542},
year={2024}
}
@article{shen2024tacogfn,
title={TacoGFN: Target Conditioned GFlowNet for Structure-Based Drug Design},
author={Shen, Tony and Seo, Seonghwan and Lee, Grayson and Pandey, Mohit and Smith, Jason R and Cherkasov, Artem and Kim, Woo Youn and Ester, Martin},
journal={arXiv preprint arXiv:2310.03223},
year={2024},
note={Published in Transactions on Machine Learning Research(TMLR)}
}
@article{seo2023molecular,
title={Molecular generative model via retrosynthetically prepared chemical building block assembly},
author={Seo, Seonghwan and Lim, Jaechang and Kim, Woo Youn},
journal={Advanced Science},
volume={10},
number={8},
pages={2206674},
year={2023},
publisher={Wiley Online Library}
}
- GFlowNet (github: recursionpharma/gflownet)
- TacoGFN [github: tsa87/TacoGFN-SBDD]
- PharmacoNet [github: SeonghwanSeo/PharmacoNet]
- UniDock [github: dptech-corp/Uni-Dock]