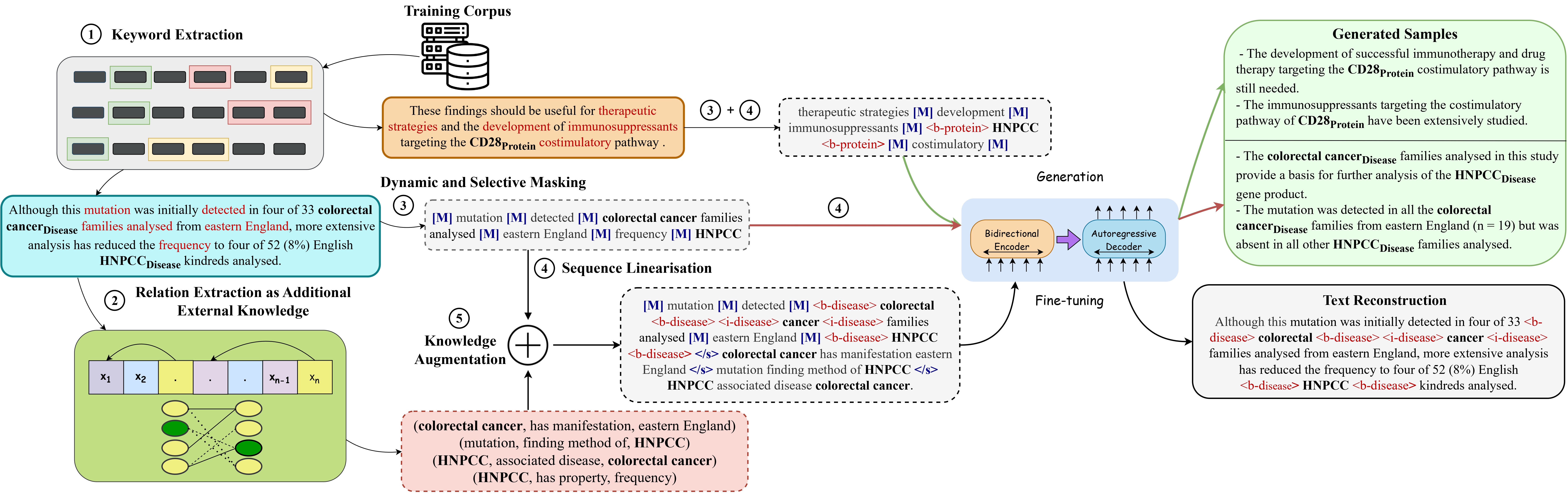
Implementation of BioAug: Conditional Generation based Data Augmentation for Low-Resource Biomedical NER
- Install dependencies using:
pip install -r requirements.txt
- Identify keywords and prepare your dataset in the format given in sample-dataset. We have uploaded the original datasets used in our paper in the datasets folder. We have also uploaded the datasets after keyword identification in the spacy-datasets folder.
Given below is the required format after keyword identification:
Word \tabspace Label \tabspace Word Class (NOUN, PRONOUN, ...) \tabspace IsKeyword? (use BIO tagging scheme)
-
Setup OpenNRE and download the checkpoint here. If you want to train your own OpenNRE model, follow the steps below:
- Prepare a dataset using this GitHub link or any other relation extraction (RE) dataset. You would need to follow Step 1 and Step 2. For Step 1, you can use the dataset here
- OpenNRE does not support pretraining random models. We had to update their code. You can use pretrain.py to update their base code. You'll also need a relation-to-id json file to train. We have provided the file for the UMLS dataset umls-rel2id.json.
-
With the help of the pretrained OpenNRE model, run preprocess.py to convert the data into the correct format and also precompute the relations between entities. (Do update lines 75, 100 and 120 according to your dataset)
Input -> Train and Dev files generated in Step 2
Output -> {train/dev}_processed.txt, {train/dev}_processed.json and {train/dev}_processed_precompute.json generated in the same directory
python preprocess.py \
--type train \
--input_file ./spacy-datasets/bc2gm/100/train.txt
We have uploaded our preprocessed files in the datasets-precompute folder.
- Run train_dynamic.sh to train your model using the following command:
sh train_dynamic.sh \
<size (100/200/500/1000/all)> \
<dataset (ebmnlp/bc5dr/bc2gm/jnlpba/ncbi)> \
<flair_batch_size> \
<SEED> \
<generations>
Example:
sh train_dynamic.sh 500 ebmnlp 8 42 5
The above step will train and generate data augmentations using BioAug followed by training a NER model on gold + augmentations using flair
Note: You'll need to replace the encoder and decoder embeddings in pretrain_dynamic.py line 246-300 if using an external dataset. Also update the new tokens in pretrain_dynamic.py and test-dynamic.py
Please cite our work:
@misc{ghosh2023bioaug,
title={BioAug: Conditional Generation based Data Augmentation for Low-Resource Biomedical NER},
author={Sreyan Ghosh and Utkarsh Tyagi and Sonal Kumar and Dinesh Manocha},
year={2023},
eprint={2305.10647},
archivePrefix={arXiv},
primaryClass={cs.CL}
}