Key Features • Installation • Usage • Training • Inference • Examples • Roadmap • Citation
Update 08.08.2023:
‼️ We fixed a severe training bug and uploaded new checkpoints. Please make sure to pull all changes and redownload your CellViT checkpoints to get the best results‼️ ☑️ Improved reproducability by providing config and log files for best models (CellViT-SAM-H and CellViT-256) and adopted PanNuke inference script for an easier evaluation
☑️ Inference speed improved by x100 for postprocessing, added new preprocessing with CuCIM speedup
☑️ Fixed bug in postprocessing that may insert doubled cells during cell-detection
☑️ Added batch-size and mixed-precision options to inference cli to support RAM limited GPUs
☑️ Extended configuration and added sweep configuration
Hörst, F., Rempe, M., Heine, L., Seibold, C., Keyl, J., Baldini, G., Ugurel, S., Siveke, J., Grünwald, B., Egger, J., & Kleesiek, J. (2023). CellViT: Vision Transformers for precise cell segmentation and classification. https://doi.org/10.48550/ARXIV.2306.15350
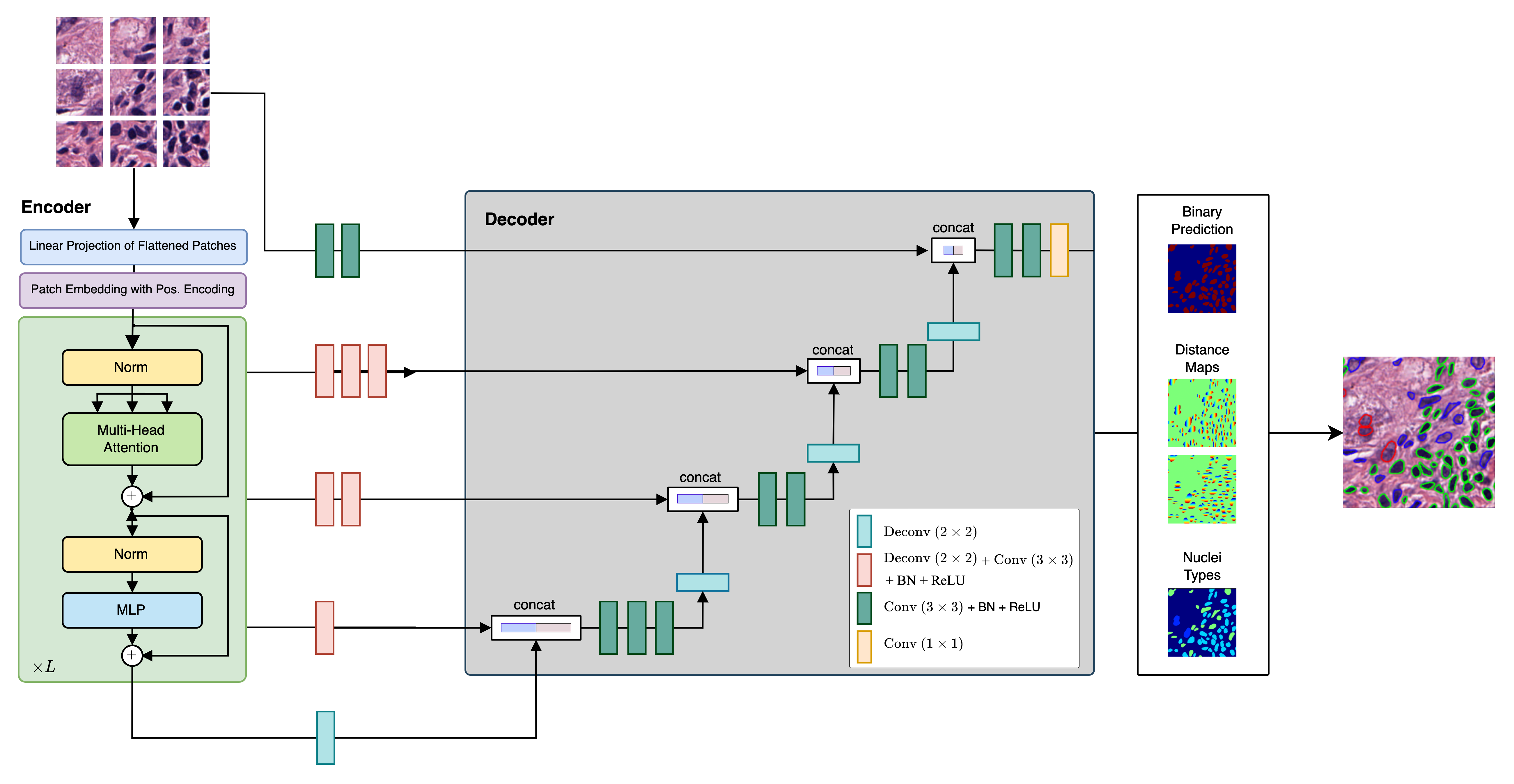
This repository contains the code implementation of CellViT, a deep learning-based method for automated instance segmentation of cell nuclei in digitized tissue samples. CellViT utilizes a Vision Transformer architecture and achieves state-of-the-art performance on the PanNuke dataset, a challenging nuclei instance segmentation benchmark.
If you intend to use anything from this repo, citation of the original publication given above is necessary
- State-of-the-Art Performance: CellViT outperforms existing methods for nuclei instance segmentation by a substantial margin, delivering superior results on the PanNuke dataset:
- Mean panoptic quality: 0.51
- F1-detection score: 0.83
- Vision Transformer Encoder: The project incorporates pre-trained Vision Transformer (ViT) encoders, which are known for their effectiveness in various computer vision tasks. This choice enhances the segmentation performance of CellViT.
- U-Net Architecture: CellViT adopts a U-Net-shaped encoder-decoder network structure, allowing for efficient and accurate nuclei instance segmentation. The network architecture facilitates both high-level and low-level feature extraction for improved segmentation results.
- Weighted Sampling Strategy: To enhance the performance of CellViT, a novel weighted sampling strategy is introduced. This strategy improves the representation of challenging nuclei instances, leading to more accurate segmentation results.
- Fast Inference on Gigapixel WSI: The framework provides fast inference results by utilizing a large inference patch size of
$1024 \times 1024$ pixels, in contrast to the conventional$256$ -pixel-sized patches. This approach enables efficient analysis of Gigapixel Whole Slide Images (WSI) and generates localizable deep features that hold potential value for downstream tasks. We provide a fast inference pipeline with connection to current Viewing Software such as QuPath
-
Clone the repository:
git clone https://github.com/TIO-IKIM/CellViT.git -
Create a conda environment with Python 3.9.7 version and install conda requirements:
conda env create -f environment.yml. You can change the environment name by editing thenametag in the environment.yaml file. This step is necessary, as we need to installOpenslidewith binary files. This is easier with conda. Otherwise, installation from source needs to be performed and packages installed with pi -
Activate environment:
conda activate cellvit_env -
Install torch (>=2.0) for your system, as described here. Preferred version is 2.0, see optional_dependencies for help. You can find all version here: https://pytorch.org/get-started/previous-versions/
-
Install optional dependencies
pip install -r optional_dependencies.txtto get a speedup using NVIDIA-Clara and CuCIM for preprocessing during inference. Please select your CUDA versions. Help for installing cucim can be found online. Note Error: cannot import name CuImage from cucim If you get this error, install cucim from conda to get all binary files. First remove your previous dependeny withpip uninstall cupy-cuda117and reinstall withconda install -c rapidsai cuciminside your conda environment. This process is time consuming, so you should be patient. Also follow their official guideline.
ResolvePackageNotFound: -gcc
- Fix: Comment out the gcc package in the environment.yml file
ResolvePackageNotFound: -libtiff==4.5.0=h6adf6a1_2, -openslide==3.4.1=h7773abc_6
- Fix: Remove the version hash from environment.yml file, such that:
... dependencies: ... - libtiff=4.5.0 - openslide=3.4.1 pip: ...
PyDantic Validation Errors for the CLI
Please install the pydantic version specified (pydantic==1.10.4), otherwise validation errors could occur for the CLI.
We are currently using the following folder structure:
├── base_ml # Basic Machine Learning Code: CLI, Trainer, Experiment, ...
├── cell_segmentation # Cell Segmentation training and inference files
│ ├── datasets # Datasets (PyTorch)
│ ├── experiments # Specific Experiment Code for different experiments
│ ├── inference # Inference code for experiment statistics and plots
│ ├── trainer # Trainer functions to train networks
│ ├── utils # Utils code
│ └── run_xxx.py # Run file to start an experiment
├── configs # Config files
│ ├── examples # Example config files with explanations
│ └── python # Python configuration file for global Python settings
├── datamodel # Datamodels of WSI, Patientes etc. (not ML specific)
├── docs # Documentation files (in addition to this main README.md)
├── models # Machine Learning Models (PyTorch implementations)
│ ├── encoders # Encoder networks (see ML structure below)
│ ├── pretrained # Checkpoint of important pretrained models (needs to be downloaded from Google drive)
│ └── segmentation # CellViT Code
├── preprocessing # Preprocessing code
│ └── patch_extraction # Code to extract patches from WSIThe CLI for a ML-experiment to train the CellViT-Network is as follows (here the run_cellvit.py script is used):
usage: run_cellvit.py [-h] --config CONFIG [--gpu GPU] [--sweep | --agent AGENT | --checkpoint CHECKPOINT]
Start an experiment with given configuration file.
optional arguments:
-h, --help show this help message and exit
--gpu GPU Cuda-GPU ID (default: None)
--sweep Starting a sweep. For this the configuration file must be structured according to WandB sweeping. Compare
https://docs.wandb.ai/guides/sweeps and https://community.wandb.ai/t/nested-sweep-configuration/3369/3 for further
information. This parameter cannot be set in the config file! (default: False)
--agent AGENT Add a new agent to the sweep. Please pass the sweep ID as argument in the way entity/project/sweep_id, e.g.,
user1/test_project/v4hwbijh. The agent configuration can be found in the WandB dashboard for the running sweep in
the sweep overview tab under launch agent. Just paste the entity/project/sweep_id given there. The provided config
file must be a sweep config file.This parameter cannot be set in the config file! (default: None)
--checkpoint CHECKPOINT
Path to a PyTorch checkpoint file. The file is loaded and continued to train with the provided settings. If this is
passed, no sweeps are possible. This parameter cannot be set in the config file! (default: None)
required named arguments:
--config CONFIG Path to a config file (default: None)The important file is the configuration file, in which all paths are set, the model configuration is given and the hyperparameters or sweeps are defined. For each specific run file, there exists an example file in the ./configs/examples/cell_segmentation folder with the same naming as well as a configuration file that explains how to run WandB sweeps for hyperparameter search. All metrics defined in your trainer are logged to WandB. The WandB configuration needs to be set up in the configuration file, but also turned off by the user.
An example config file is given here with explanations here.
For sweeps, we provide a sweep example file train_cellvit_sweep.yaml.
Pre-trained ViT models for training initialization can be downloaded from Google Drive: ViT-Models. Please check out the corresponding licenses before distribution and further usage! Note: We just used the teacher models for ViT-256.
❗ If your training crashes at some point, you can continue from a checkpoint
We use a customized dataset structure for the PanNuke and the MoNuSeg dataset.
The dataset structures are explained in pannuke.md and monuseg.md documentation files.
We also provide preparation scripts in the cell_segmentation/datasets/ folder.
In our paper, we did not (!) use early stopping, but rather train all models for 130 to eliminate selection bias but have the largest possible database for training. Therefore, evaluation neeeds to be performed with the latest_checkpoint.pth model and not the best early stopping model.
We provide to script to create evaluation results: inference_cellvit_experiment.py for PanNuke and inference_cellvit_monuseg.py for MoNuSeg.
❗ We recently adapted the evaluation code and added a tag to the config files to select which checkpoint needs to be used. Please make sure to use the right checkpoint and select the appropriate dataset magnification.
Model checkpoints can be downloaded here:
License: Apache 2.0 with Commons Clause
Proved checkpoints have been trained on 90% of the data from all folds with the settings described in the publication.
The following steps are necessary for preprocessing:
- Prepare WSI with our preprocessing pipeline
- Run inference with the
inference/cell_detection.pyscript
Results are stored at preprocessing locations
In our Pre-Processing pipeline, we are able to extract quadratic patches from detected tissue areas, load annotation files (.json) and apply color normlizations. We make use of the popular OpenSlide library, but extended it with the RAPIDS cuCIM framework for an x8 speedup in patch-extraction. The documentation for the preprocessing can be found here.
Preprocessing is necessary to extract patches for our inference pipeline. We use squared patches of size 1024 pixels with an overlap of 64 px.
Please make sure that you select the following properties for our CellViT inference
| Parameter | Value |
|---|---|
| patch_size | 1024 |
| patch_overlap | 6.25 |
In general, the folder structure for a preprocessed dataset looks like this: The aim of pre-processing is to create one dataset per WSI in the following structure:
WSI_Name
├── annotation_masks # thumbnails of extracted annotation masks
│ ├── all_overlaid.png # all with same dimension as the thumbnail
│ ├── tumor.png
│ └── ...
├── context # context patches, if extracted
│ ├── 2 # subfolder for each scale
│ │ ├── WSI_Name_row1_col1_context_2.png
│ │ ├── WSI_Name_row2_col1_context_2.png
│ │ └── ...
│ └── 4
│ │ ├── WSI_Name_row1_col1_context_2.png
│ │ ├── WSI_Name_row2_col1_context_2.png
│ │ └── ...
├── masks # Mask (numpy) files for each patch -> optional folder for segmentation
│ ├── WSI_Name_row1_col1.npy
│ ├── WSI_Name_row2_col1.npy
│ └── ...
├── metadata # Metadata files for each patch
│ ├── WSI_Name_row1_col1.yaml
│ ├── WSI_Name_row2_col1.yaml
│ └── ...
├── patches # Patches as .png files
│ ├── WSI_Name_row1_col1.png
│ ├── WSI_Name_row2_col1.png
│ └── ...
├── thumbnails # Different kind of thumbnails
│ ├── thumbnail_mpp_5.png
│ ├── thumbnail_downsample_32.png
│ └── ...
├── tissue_masks # Tissue mask images for checking
│ ├── mask.png # all with same dimension as the thumbnail
│ ├── mask_nogrid.png
│ └── tissue_grid.png
├── mask.png # tissue mask with green grid
├── metadata.yaml # WSI metdata for patch extraction
├── patch_metadata.json # Patch metadata of WSI merged in one file
└── thumbnail.png # WSI thumbnailThe cell detection and segmentation results are stored in a newly created cell_detection folder for each WSI.
If the data is prepared, use the cell_detection.py script inside the cell_segmentation/inference folder to perform inference:
usage: cell_detection.py --model MODEL [--gpu GPU] [--magnification MAGNIFICATION] [--mixed_precision]
[--batch_size BATCH_SIZE] [--outdir_subdir OUTDIR_SUBDIR]
[--geojson] {process_wsi,process_dataset} ...
Perform CellViT inference for given run-directory with model checkpoints and logs
optional arguments:
-h, --help show this help message and exit
--gpu GPU Cuda-GPU ID for inference. Default: 0 (default: 0)
--magnification MAGNIFICATION
Network magnification. Is used for checking patch magnification such that
we use the correct resolution for network. Default: 40 (default: 40)
--mixed_precision Whether to use mixed precision for inference. Default: False (default: False)
--batch_size BATCH_SIZE
Inference batch-size. Default: 8 (default: 8)
--outdir_subdir OUTDIR_SUBDIR
If provided, a subdir with the given name is created in the cell_detection folder
where the results are stored. Default: None (default: None)
--geojson Set this flag to export results as additional geojson files for
loading them into Software like QuPath. (default: False)
required named arguments:
--model MODEL Model checkpoint file that is used for inference (default: None)
subcommands:
Main run command for either performing inference on single WSI-file or on whole dataset
{process_wsi,process_dataset}For processing a single WSI file, you need to select the process_wsi (python3 cell_detection.py process_wsi) subcommand with the following structure:
usage: cell_detection.py process_wsi --wsi_path WSI_PATH --patched_slide_path PATCHED_SLIDE_PATH
Process a single WSI file
arguments:
-h, --help show this help message and exit
--wsi_path WSI_PATH Path to WSI file
--patched_slide_path PATCHED_SLIDE_PATH
Path to patched WSI file (specific WSI file, not parent path of patched slide dataset)To process an entire dataset, select process_dataset (python3 cell_detection.py process_dataset):
usage: cell_detection.py process_dataset --wsi_paths WSI_PATHS --patch_dataset_path PATCH_DATASET_PATH [--filelist FILELIST] [--wsi_extension WSI_EXTENSION]
Process a whole dataset
arguments:
-h, --help show this help message and exit
--wsi_paths WSI_PATHS
Path to the folder where all WSI are stored
--patch_dataset_path PATCH_DATASET_PATH
Path to the folder where the patch dataset is stored
--filelist FILELIST Filelist with WSI to process. Must be a .csv file with one row denoting the filenames (named 'Filename').
If not provided, all WSI files with given ending in the WSI folder are processed. (default: 'None')
--wsi_extension WSI_EXTENSION
The extension types used for the WSI files, see configs.python.config (WSI_EXT). (default: 'svs')We provide an example TCGA file to show the performance and usage of our algorithms. Files and scripts can be found in the example folder. The TCGA slide must be downloaded here: https://portal.gdc.cancer.gov/files/f9147f06-2902-4a64-b293-5dbf9217c668. Please place this file in the example folder.
Preprocessing:
python3 ./preprocessing/patch_extraction/main_extraction.py --config ./example/preprocessing_example.yamlOutput is stored inside ./example/output/preprocessing
Inference:
Download the models and store them in ./models/pretrained or on your preferred location and change the model parameter.
python3 ./cell_segmentation/inference/cell_detection.py \
--model ./models/pretrained/CellViT/CellViT-SAM-H-x40.pth\
--gpu 0 \
--geojson \
process_wsi \
--wsi_path ./example/TCGA-V5-A7RE-11A-01-TS1.57401526-EF9E-49AC-8FF6-B4F9652311CE.svs \
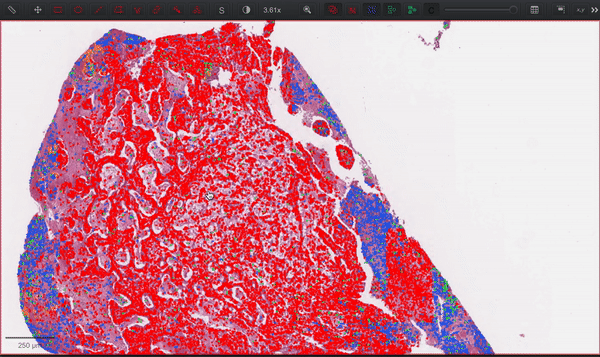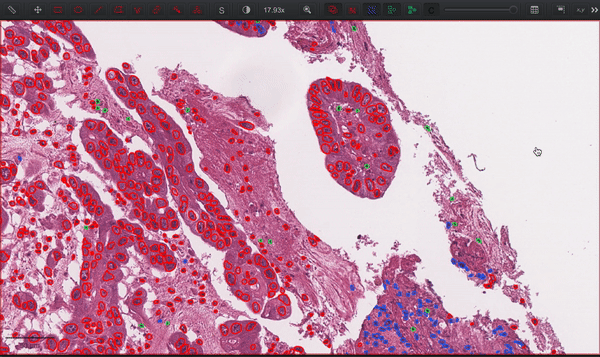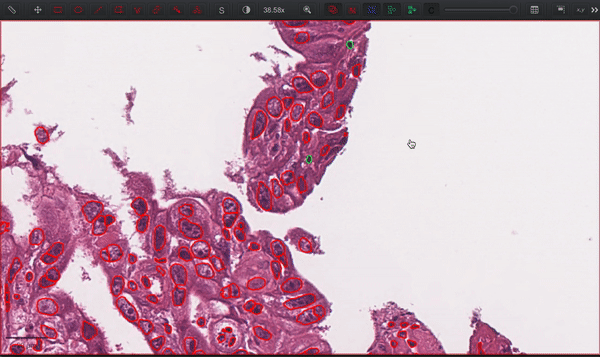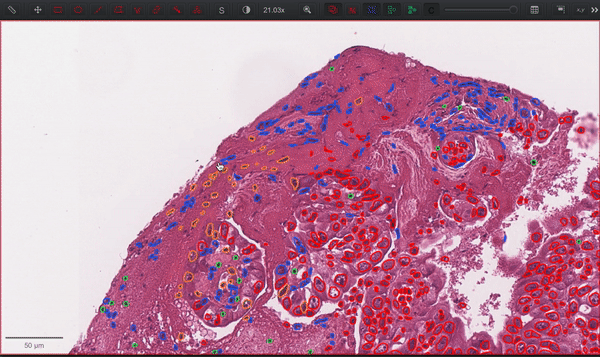
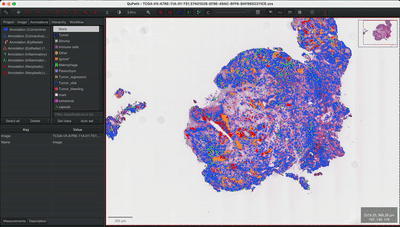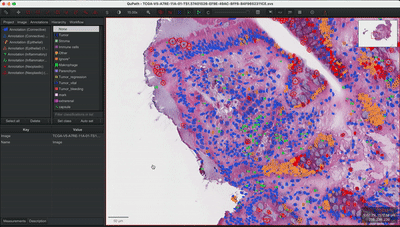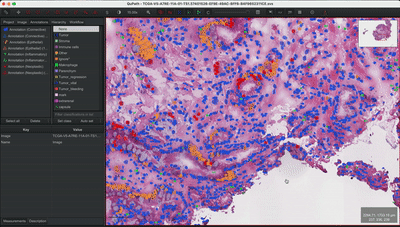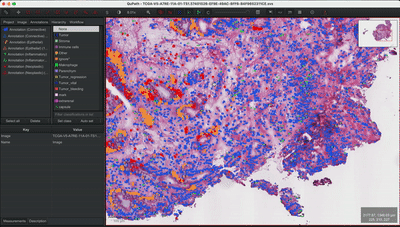
--patched_slide_path ./example/output/preprocessing/TCGA-V5-A7RE-11A-01-TS1.57401526-EF9E-49AC-8FF6-B4F9652311CEYou can import your results (.geojson files) into QuPath. The results should look like this:
We are currently optimizing the inference speed. Code will be updated in a few weeks.
In a future release, we will provide a Docker image that contains all the necessary dependencies and configurations pre-installed. This Docker image will ensure reproducibility and simplify the setup process, allowing for easy installation and usage of the project.
Stay tuned for updates on the availability of the Docker image, as we are actively working on providing this convenient packaging option for our project. 🚀
@article{CellViT,
title = {CellViT: Vision Transformers for precise cell segmentation and classification},
journal = {Medical Image Analysis},
volume = {94},
pages = {103143},
year = {2024},
issn = {1361-8415},
doi = {https://doi.org/10.1016/j.media.2024.103143},
url = {https://www.sciencedirect.com/science/article/pii/S1361841524000689},
author = {Fabian Hörst and Moritz Rempe and Lukas Heine and Constantin Seibold and Julius Keyl and Giulia Baldini and Selma Ugurel and Jens Siveke and Barbara Grünwald and Jan Egger and Jens Kleesiek},
}