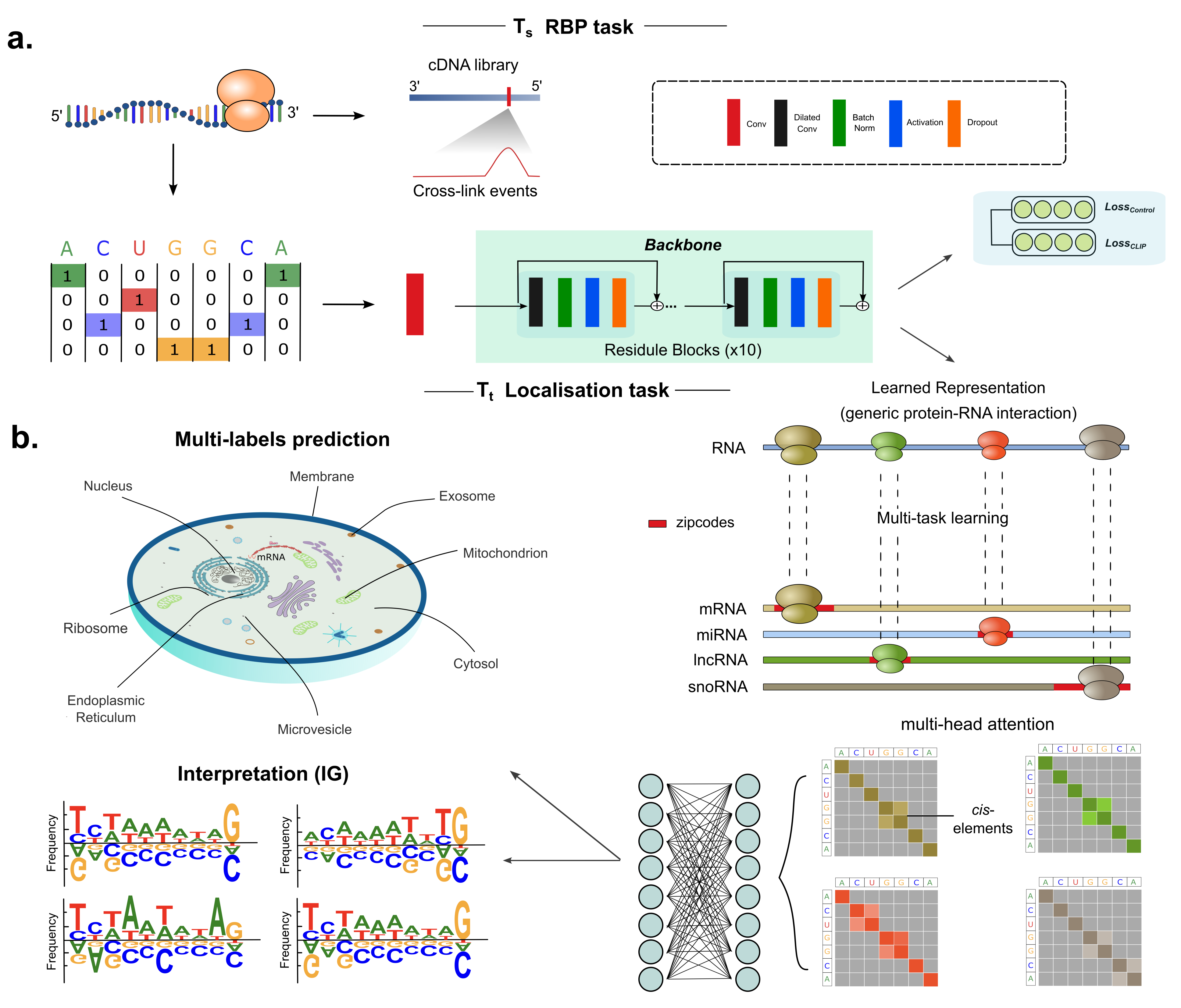
DeeplocRNA is a deep neural network that predicts the RNA localization, enabling the prediction across 4 RNA types (mRNA, miRNA, lncRNA, snoRNA) and different species (Human and Mouse).
Please make sure anaconda is installed in your local machine, create a new working environment to run DeepLocRNA
conda create -n DeepLocRNA python=3.8
Enter the new created environment
source activate DeepLocRNA
To run the model, you should download DeepLocRNA via git or pypi
From git
pip install git+https://github.com/TerminatorJ/DeepLocRNA.git
or from pypi
pip install DeepLocRNA
Install two dependenies
pip install tensorflow==2.4.1
pip install typing-extensions==4.7.1
if you want to train the model youself, please follow the following steps
DeepLocRNA works on FASTA files, e.g.
>test1 ACTGCCGTATCGTAGCTAGCTAGTGATCGTAGCTACGTAGCTAGCTAGCTACGATCGTAGTCAGTCGTAGTACGTCA >test2 ACACACATGAGCGATGTAGTCGATGATGCATCGACGATCGATCGAGCTACGTAGCATCGATCGATGCATCGACGTAG
One can aldo use our prepared dataset to train the model as below
wget https://github.com/TerminatorJ/DeepLocRNA/archive/refs/heads/main.zip
Then compress the zip file
unzip main.zip
cd ./DeepLocRNA-main/DeepLocRNA
python ./fine_tuning_deeprbploc_allRNA.py --dataset ./data/allRNA/allRNA_all_human_data_seq_mergedm3locall2_deduplicated2_filtermilnc.fasta
Afterwards, there will be "*_X.npy" in the "./DeepLocRNA/data/allRNA/allRNA_all_human_data_seq_mergedm3locall2_deduplicated2_filtermilnc" folder.
In order to do multiple RNA prediction, we will generate tags for all RNA species
python ./fine_tuning_deeprbploc_allRNA.py --dataset ./data/allRNA/allRNA_all_human_data_seq_mergedm3locall2_deduplicated2_filtermilnc.fasta --RNA_tag
Afterwards, there will be "*_X_tag.npy" in the "./DeepLocRNA/data/allRNA/allRNA_all_human_data_seq_mergedm3locall2_deduplicated2_filtermilnc" folder.
To train the model locally, RBP pre-trained model should be loaded first.
pip install ../parnet-develop
We provide two options to train the model
First, you can use standard training strategy, using single GPU to train the model. It is worth note that the training is bonded with 5-folds as default, which will repeat 5 times to go through the data.
python ./fine_tuning_deeprbploc_allRNA.py --dataset ./data/allRNA/allRNA_all_human_data_seq_mergedm3locall2_deduplicated2_filtermilnc.fasta --load_data --gpu_num 1 --species human --batch_size 8 --flatten_tag --gradient_clip --loss_type BCE --jobnum 001 --species Human
Arguments for training
| Long | Description |
|---|---|
--dataset |
Path to the input fasta file, which is a mixture of different RNA types. Formatted according to the input format. |
--load_data |
loading the saved data, you should add this argument after generating X.npy and X_tag.npy. |
--gpu_num |
The number of gpus you want to use while training the model. |
--species |
The species you want to predict |
--batch_size |
The batch size while training the model for each step. |
--flatten_tag |
Add this tag to enable multi-RNA training. |
--gradient_clip |
whether using gradient clip to make the training process stable. |
--loss_type |
The loss function that used to train the model. Default: BCE. |
--jobnum |
The identified number that represents the run of each training job. |
There should prepare your input file to ".fasta"(#Input-format) format
python ./DeepLocRNA/fine_tuning_deeprbploc_allRNA_prediction.py --fasta ./example.fasta --rna_types mRNA --species Human
Alternatively, you can also use our online webserver if you only have a couple sequences to be predicted ()
python fine_tuning_deeprbploc_allRNA_prediction.py --fasta ./example.fasta --rna_types mRNA --species Human --plot True
If you wish to get the precise nucleotide contribution, please choose "--plot" as True, and define the configure file yourself as "att_config.csv" before input in the input frame.
starts,ends
10,100
50,100
Where 10 to 100 is the interval that you want to get the attribution scores.
python fine_tuning_deeprbploc_allRNA_prediction.py --fasta ./example.fasta --rna_types lncRNA --att_config att_config.csv --species Human --plot True
Arguments for prediction
| Long | Description |
|---|---|
--fasta |
Path to the input fasta file, formatted according to the input format. |
--rna_types |
The type of RNA you want to predict |
--species |
The species you want to predict |
--att_config |
Path to the customized position of a specific sequence as .csv. formatted according to att config format |
--plot |
Plot the attribution figures, this is mandatory if you want to visualize the explaination plots. Default: True |