Arash Hajisafi, Haowen Lin, Yao-Yi Chiang, and Cyrus Shahabi. "Dynamic GNNs for Precise Seizure Detection and Classification from EEG Data." In Pacific-Asia Conference on Knowledge Discovery and Data Mining, pp. 207-220. Singapore: Springer Nature Singapore, 2024. https://arxiv.org/abs/2405.09568
This repository hosts the implementation of NeuroGNN, a dynamic Graph Neural Network (GNN) framework introduced in our paper "Dynamic GNNs for Precise Seizure Detection and Classification from EEG Data", accepted at PAKDD '24. Our framework is designed to enhance seizure detection and classification through the capture of EEG data's spatial, temporal, semantic, and taxonomic correlations.
NeuroGNN captures the dynamic interplay between EEG electrode locations and the semantics of their corresponding brain regions, leveraging the intricate relationships governed by each brain region's distinct cognitive functions and sensory processing. This multifaceted approach allows for a comprehensive understanding of brain activity, leading to improved precision in seizure detection and classification.
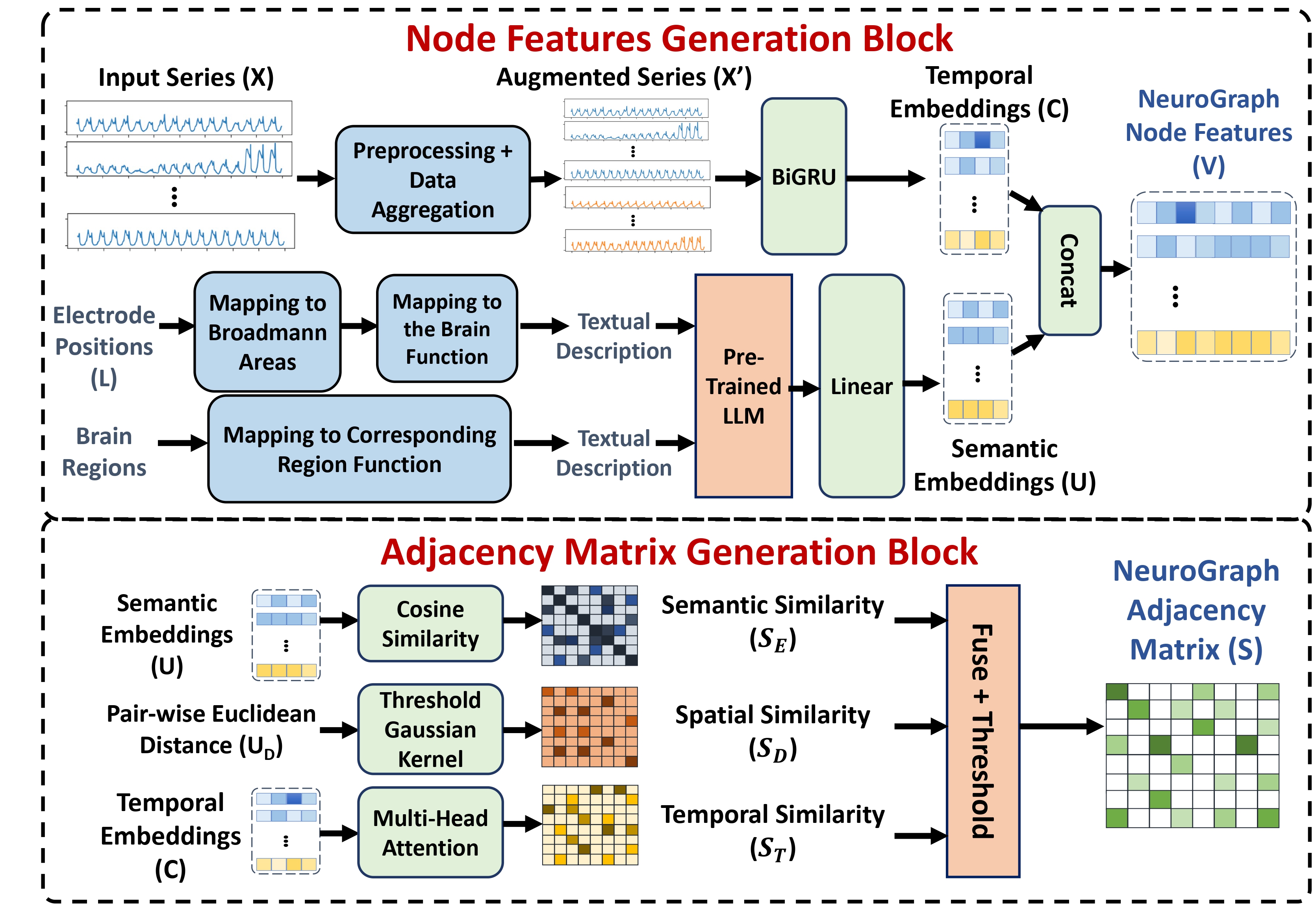
Figure 1: NeuroGNN Graph Construction - Fusing spatial, temporal, semantic, and taxonomic correlations.
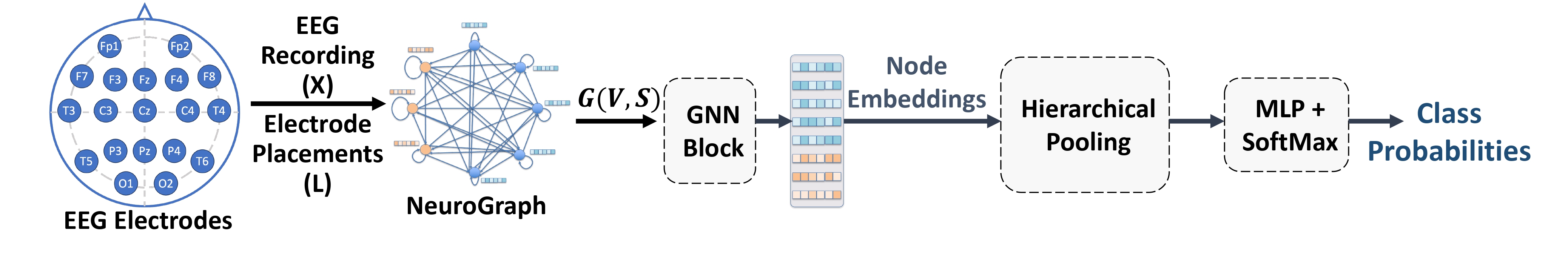
Figure 2: Seizure Detection & Classification Using NeuroGNN.
The constants.py file contains important descriptions and mappings, including the textual descriptions for brain regions and Brodmann areas used to generate semantic embeddings, and the mapping from EEG electrodes to these areas and regions. Our paper provides further details on how these elements are utilized within the NeuroGNN framework.
This project is forked from the repository of "Self-Supervised Graph Neural Networks for Improved Electroencephalographic Seizure Analysis" by Siyi Tang et al. We reuse portions of their code, including pre-processing scripts, training loops, and evaluation codes. Additionally, the sections of this documentation related to dataset preparation and preprocessing are based on their comprehensive documentation. Check out their original work, licensed under the MIT License.
To install the required dependencies for NeuroGNN, follow these steps:
- Clone the NeuroGNN repository
git clone https://github.com/USC-InfoLab/NeuroGNN.git- Navigate to the NeuroGNN directory
cd NeuroGNN- Install the required Python packages
pip install -r requirements.txtWe use the Temple University Seizure Corpus (TUSZ) v1.5.2 in this study. The TUSZ dataset can be accessed through here, where you need to sign up through the official data provider's website. After you have registered and downloaded the data, you will see a subdirectory called edf which contains all the EEG signals and their associated labels. We use the EEG files in the edf/dev subfolder as our held-out test set. We further split the EEG files in the edf/train subfolder into train and validation sets by patients. See folders ./data/file_markers_detection, ./data/file_markers_classification, and ./data/file_markers_ssl for details.
In this study, we exclude five patients from the test set who exist in both the official TUSZ train and test sets. You can find the list of excluded patients' IDs in ./data_tusz/excluded_test_patients.txt.
In addition, ./data_tusz/focal_labeled_as_generalized.csv provides the list of 27 seizures in the test set that we think are focal seizures (manually analyzed by a board-certified EEG reader) but are labeled as generalized non-specific seizures in TUSZ data. See the paper by Tang et al. for more details.
The preprocessing step resamples all EEG signals to 200Hz, and saves the resampled signals in 19 EEG channels as h5 files.
On terminal, run the following:
python ./data/resample_signals.py --raw_edf_dir <tusz-data-dir> --save_dir <resampled-dir>where <tusz-data-dir> is the directory where the downloaded TUSZ v1.5.2 data are located, and <resampled-dir> is the directory where the resampled signals will be saved.
Note that the remaining preprocessing step in our paper (Fourier transform on short sliding windows) is handled by dataloaders. You can (optionally) perform this preprocessing step prior to model training to accelerate the training.
Preprocessing for seizure detection and self-supervised pre-training:
python ./data/preprocess_detection.py --resampled_dir <resampled-dir> --raw_data_dir <tusz-data-dir> --output_dir <preproc-dir> --clip_len <clip-len> --time_step_size 1 --is_fftwhere <clip-len> is 60 or 12.
Preprocessing for seizure classification:
python ./data/preprocess_classification.py --resampled_dir <resampled-dir> --raw_data_dir <tusz-data-dir> --output_dir <preproc-dir> --clip_len <clip-len> --time_step_size 1 --is_fftFor those interested in tracking and visualizing their training experiments, you can optionally log your results to Weights & Biases. If you don't have a wandb account, you can create one at wandb.ai. To enable wandb logging, add the --use_wandb flag to your training commands. This is entirely optional and can be omitted if you prefer not to use wandb.
To train seizure detection from scratch using NeuroGNN, run:
python train.py --input_dir <resampled-dir> --raw_data_dir <tusz-data-dir> --save_dir <save-dir> --max_seq_len <clip-len> --do_train --num_epochs 100 --task detection --metric_name auroc --use_fft --lr_init 1e-4 --num_classes 1 --data_augment --model_name neurognnwhere <clip-len> is 60 or 12.
To use preprocessed Fourier transformed inputs from the above optional preprocessing step, specify --preproc_dir <preproc-dir>.
To train seizure type classification from scratch using NeuroGNN, run:
python train.py --input_dir <resampled-dir> --raw_data_dir <tusz-data-dir> --save_dir <save-dir> --max_seq_len <clip-len> --do_train --num_epochs 60 --task classification --metric_name F1 --use_fft --lr_init 2e-4 --num_classes 4 --data_augment --dropout 0.5 --model_name neurognnSimilarly, <clip-len> is 60 or 12. To use preprocessed Fourier transformed inputs from the above optional preprocessing step, specify --preproc_dir <preproc-dir>.
To train self-supervised next time period prediction using NeuroGNN, run:
python train_ssl.py --input_dir <resampled-dir> --raw_data_dir <tusz-data-dir> --save_dir <save-dir> --max_seq_len <clip-len> --output_seq_len 12 --do_train --num_epochs 350 --task 'SS pre-training' --metric_name loss --use_fft --lr_init 5e-4 --data_augment --model_name neurognnSimilarly, <clip-len> is 60 or 12. To use preprocessed Fourier transformed inputs from the above optional preprocessing step, specify --preproc_dir <preproc-dir>.
To fine-tune seizure detection/seizure type classification models from self-supervised pre-training, add the following additional arguments:
--fine_tune --load_model_path <pretrained-model-checkpoint>We provide pretrained checkpoints for seizure detection, classification, and self-supervised learning tasks. These models are available in the neurognn_checkpoints directory of our repository. You can leverage these models to kickstart your research or applications related to EEG data analysis.
If you find our work useful, please consider citing:
@inproceedings{hajisafi2024dynamic,
title = {Dynamic GNNs for Precise Seizure Detection and Classification from EEG Data},
author = {Hajisafi, Arash and Lin, Haowen and Chiang, Yao-Yi and Shahabi, Cyrus},
booktitle = {Pacific-Asia Conference on Knowledge Discovery and Data Mining},
pages = {207--220},
year = {2024},
organization = {Springer}
}
This project is licensed under the MIT License - see the LICENSE file for details.
