This repository contains the course materials for hands-on exercise of the "Variants Annotation and Phenotype Analysis" session in the Quantitative Genomics workshop at Columbia University on June 11-12, 2020.
Both the lectures and hands-on exercises will be taught via Zoom video conference online. To ensure the cross-platform compatibility, we will only use tools that are developed in Perl and Python. As most users are using either Windows or MacOS in your personal computers, below we describe the preferred means to prepare computing environment using Conda.
For some users, Conda is not strictly required if you are already familiar with running command-line driven software tools in Windows via WSL or PowerShell and have Perl/Python pre-installed in your Windows computer. So if you are among this group of users, you can skip this step and go directly to the exercises.
For other users, the suggested way to perform the hands-on exercises is to install Anaconda, which is an open source, flexible solution that provides the utilities to build, distribute, install, update, and manage software in a cross-platform manner. Note that the Windows versions of several widely graphical user interface (GUI) packages, including Jupyter Notebook and Spyder, are already included within Anaconda by default; additionally, Rstudio can be also installed by one-click installation within Anaconda Navigator.
Once you go to the website, click "Download" button (see image below), and then install in your Windows computer by accepting all default options.
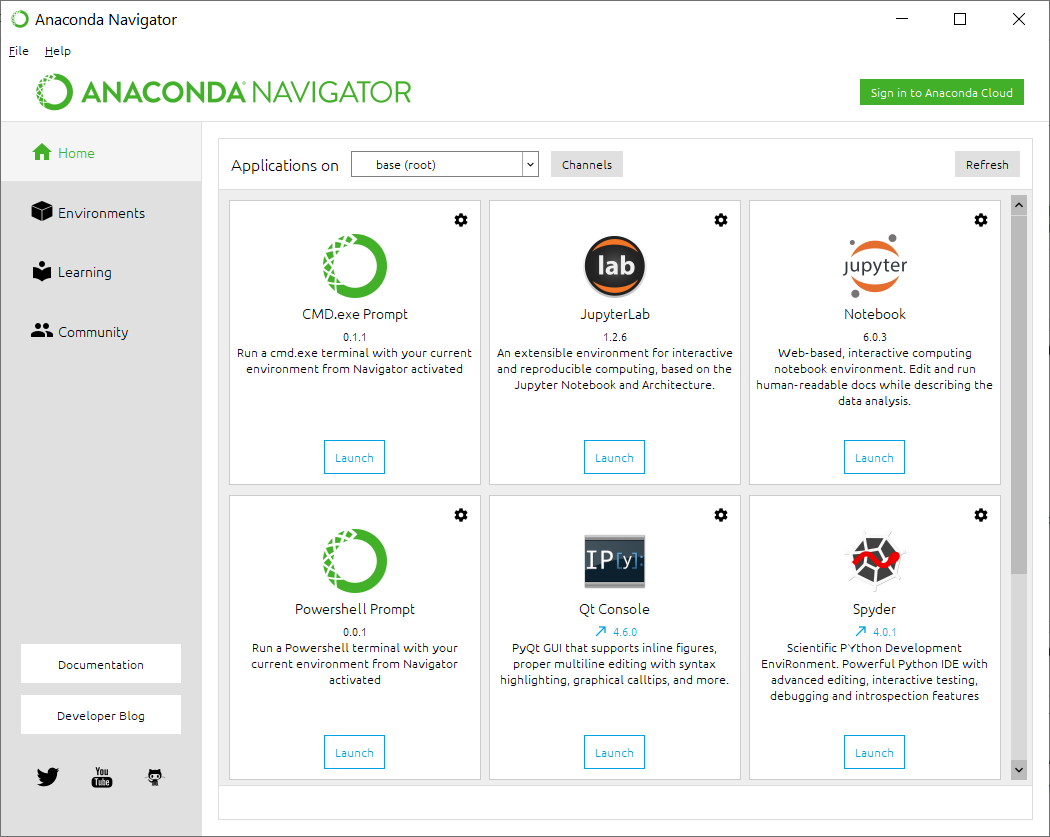
Once installation is successful, you can click the lower left "Start" button in Windows, then go to "Anaconda3" menu, then launch the "Anaconda Navigator". You will see the interface below.
Since MacOS has built-in terminal and Linux-like enrivonment for executing command-line software tools, in general you do not need any specific set up. However, you may want to install Miniconda to help manage dependencies and the computing environment. Basically you first download the Miniconda installer for macOS, then run bash Miniconda3-latest-MacOSX-x86_64.sh. Miniconda is a light version of Conda.
If you use Mac or Linux operating system, you already have terminal access and can run all basic Linux commands. If you use Windows, after installing Anaconda, you will interact with a Linux-like environment. If you are not familiar with basic Linux commands, you can follow one simple tutorial to learn several commands: https://maker.pro/linux/tutorial/basic-linux-commands-for-beginners. Some of the commands that we will use in the exercise include ls, pwd, cd, mv and wget.
If you are using Mac, not that Perl is already included as part of the MacOS. You can open a terminal window, and type perl -v to double check this. So you can skip this section.
If you are using Windows, note that Perl interpreter is not installed by default in most Windows computers. The annotation software (ANNOVAR) that we will use in this exercise is written in Perl, so we need to install an interpreter. This is where Conda can become very convenient. Within the Anaconda Navigator, click "Powershell prompt", and you will see a terminal that has "(base) C:\Users\Kai Wang>" as the prompt.
By default, we start in a base environment within the Windows Powershell. For the exercise on variants annotation, we will create an environment called annotation to use for the variants annotation and phenotype analysis.
(base) C:\Users\Kai Wang>conda create --name annotation
Next, we will enter the new environment that was just created to perform the exercise:
(base) C:\Users\Kai Wang>conda activate annotation
(annotation) C:\Users\Kai Wang>
You can see that the (base) in command prompt is changed to (annotation) indicating that we are now in a different environment.
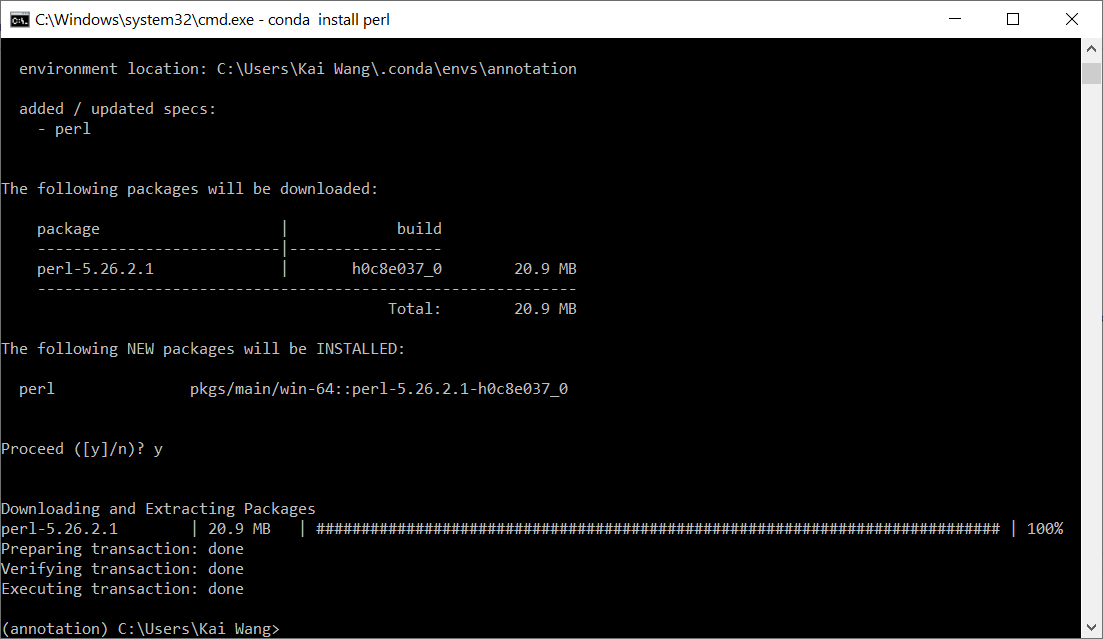
You can do conda install perl, and press "y" when prompted. See image below.
After installation, you can type perl -v to double check that Perl is indeed installed by the command above in the annotation environment. You should see a message like This is perl 5, version 26, subversion 2 (v5.26.2) built for MSWin32-x64-multi-thread.
When using Windows, it is also highly recommended that you install the gzip-related tools (not required for this exercise itself, but can be useful if you want to explore more functionalities of ANNOVAR by downloading additional annotation packages). You can do conda install -c msys2 m2-gzip to install gzip.
Now you can try switch back to the base environment (type conda activate base to do this), and type perl -v. You will see an error message now that this command is not recognized; in other words, Perl is only available in the annotation but not base environment. So this exercise shows how Conda manages software packages in an environment-dependent manner.
Typically you will go to the ANNOVAR website, fill in a registration form, and download the package there. For this exercise, we already prepared a ZIP file that contains a "compact" version of ANNOVAR and necessary library files, to make it easier for users. If you are using Windows, make sure to switch to the annotation environment first, and by default you will be at your home directory (you can type pwd to check this, which shows the current directory you are in). To make it easier to manage files and directories, you can create a new directory, then enter this new directory (type mkdir genomics_exercise followed by cd genomics_exercise). To confirm which directory you are in, you can type pwd.
Next, you can just download the ZIP file for this class by the command wget -O exercise1.zip https://github.com/WGLab/Workshop_Annotation/releases/download/v1.0.0/exercise1.zip. The Linux command wget essentially downloads a file from a given URL and saves the file to your computer. Because this file contains several annotation databases, its size is around 500Mb and it may take a while to download it. To unzip the file, you can dirctly using tar -xvf exercise1.zip to unzip the downladed file. You will see from the messages in screen that several files are extracted from the zip file.
Type cd exercise1 to enter the exercise1 directory. The sub-folder humandb folder already contains several annotation databases for human genome that we will use in our exercise. (Note that users can find more annotation databases here.
perl table_annovar.pl example/ex2.vcf humandb/ -buildver hg19 -out myanno -remove -protocol refGeneWithVer,cytoBand,gnomad211_exome -operation g,r,f -nastring . -vcfinput -polish
After that, you will find the result files myanno.hg19_multianno.txt and myanno.hg19_multianno.vcf.
In the command above, we specified three protocols, including RefGene annotation (a gene-based annotation), cytogenetic band annotation (a region-based annotation), and allele frequency in gnoMAD version 2.1.1 (a filter-based annotation). These three types of annotation are indicated as g, r and f in the -operation argument, respectively. We also specify that dots ('.') be used to indicate lack of information, so variants that are not observed in the database will have '.' as the annotation. The -vcfinput argument specifies that the input file is in VCF format.
If you have Excel installed, you can open the hg19_multianno.txt file by Excel and examine the various tab-delimited fields.
Next, we want to download a VCF file and then run ANNOVAR on this file.
In the exercise1 folder:
wget http://molecularcasestudies.cshlp.org/content/suppl/2016/10/11/mcs.a001131.DC1/Supp_File_2_KBG_family_Utah_VCF_files.zip -O Supp_File_2_KBG_family_Utah_VCF_files.zip
Next unzip the ZIP file (unzip in Mac/Linux can work). Windows users, just unzip here, and rename folder to VCF_files:
tar -xvf Supp_File_2_KBG_family_Utah_VCF_files.zip
mv '.\File 2_KBG family Utah_VCF files\' VCF_files
Note that if you use Mac/Linux, the command should be mv './File 2_KBG family Utah_VCF files/' VCF_files
Run ANNOVAR on the VCF file:
perl table_annovar.pl VCF_files/proband.vcf -buildver hg19 humandb -out proband.annovar -remove -protocol refGeneWithVer,gnomad211_exome -operation g,f -nastring . -vcfinput
The proband.annovar.hg19_multianno.txt file contains annotations for this exome.
Suppose we sequenced a patient with COVID-19 and performed alignment, filtering, read assembly, and generate variant calls (the calls are generated by comparing to reference genome NC045512.2). We want to evaluate what effects that the virus mutations in this patient may have.
You can prepare a simple text file called ex3.avinput with the following mutations:
NC_045512v2 29095 29095 C T
NC_045512v2 26144 26144 G T
NC_045512v2 28144 28144 T C
This format is referred to as the avinput format, and it is a simple tab or space delimited file, with each variant per line. There can be many columns per line, but the first five columns in each line represent chr, start, end, reference allele and alternative allele, respectively.
Then you can just run perl table_annovar.pl example/ex3.avinput sarscov2db -build NC_045512v2 -protocol avGene -operation g -out ex3 to annotate these mutations and the results will be stored in ex3.NC_045512v2_multianno.txt.
Alternatively, since you are using only one single type of annotation (gene-based annotation), you may also do perl annotate_variation.pl example/ex3.avinput sarscov2db/ -build NC_045512v2 -dbtype avGene -out ex3 to annotate these mutations. Examine the output file ex3.exonic_variant_function to see what proteins these mutations affect, and what amino acid changes that they cause.
In the next exercise, we will use a software tool called Phen2Gene to prioritize genes based on clinical phenotypes of patients with Mendelian diseases.
You need to install Phen2Gene using the command below. Please deactivate other conda environments if you are not in conda base.
To make the tutorial easy, the users can just download the source code for Phen2Gene from the release:
wget https://github.com/WGLab/Phen2Gene/archive/1.2.2.zip -O Phen2Gene.zip
Then extract the ZIP file and also rename the folder, run the command below (note that in Linux/MacOS, you can use unzip to unzip a file):
tar -xvf Phen2Gene.zip
mv Phen2Gene-1.2.1 Phen2Gene
cd Phen2Gene
If you are more experienced, and you want to download Phen2Gene from github, you can install git using the command below (if git is not yet available in your system) and git clone the Phen2Gene repository directly.
conda install git
git clone https://github.com/WGLab/Phen2Gene.git
cd Phen2Gene
To create your environment (necessary for ALL operating systems):
conda env create -f environment.yml
conda activate phen2gene
If on MacOS or Linux run:
bash setup.sh
And the installation will be extremely easy for you. The database and all will be downloaded and you can add Phen2Gene in your path.
If on Windows, go to the lib/ directory and download the knowledgebase file:
cd lib
wget https://github.com/WGLab/Phen2Gene/releases/download/1.1.0/H2GKBs.zip -O H2GKB.zip
Note that this file is over 1GB, so it may take a while depending on your Internet speed.
Then extract the file:
tar -xvf H2GKB.zip
We recommend extracting the file into lib directory for ease of use for this tutorial, but you can put the file wherever you want.
If on Windows, and you do not have the space to download and extract the H2GKB.zip in the lib folder (recommended default) you will have to use -d full_path_to_H2GKB.zip_extraction_folder. This means wherever you decided to download and unzip H2GKB.zip, you have to add that path to each use of phen2gene.py as below:
- Input HPO IDs via input file (typical use case)
Default command:
python phen2gene.py -f example/HPO_sample.txt -out out/prioritizedgenelist
Alternate folder for H2GKB, add -d:
python phen2gene.py -f example/HPO_sample.txt -out out/prioritizedgenelist -d full_path_to_H2GKB.zip_folder
- Input HPO IDs manually, if desired
python phen2gene.py -m HP:0000021 HP:0000027 HP:0030905 HP:0010628 -out out/prioritizedgenelist
We will use the actual variants from exome sequencing on a proband with developmental disease from a CSH Molecular Case Studies paper. In the previous exercise we already generated annotation as proband.annovar.hg19_multianno.txt file.
We already have the HPO IDs for this patient in the example folder of Phen2Gene so just run:
python phen2gene.py -f example/ANKRD11_id.txt -w sk -out ankrd11
Now, we can use awk to do a simple filtering on gnomAD allele frequency (column 11, across 125,748 exomes). We want to extract rare variants (<1% freq here, though many people use much lower number).
conda install gawk
awk '$11 <= 0.01 || $11 == "."' FS="\t" proband.annovar.hg19_multianno.txt > filtered.proband.annovar.hg19_multianno.txt
Some Windows users reported that the conda install gawk above does not work; if this applies to you, you can instead do conda install m2-base which will install many Unix-like tools including gawk.
Now using a python script that we pre-wrote in the example folder, we can create a newly sorted list of genes from Phen2Gene based on the genes with rare variants present in the ANNOVAR output:
python example/filterbyannovar.py -pre ankrd11/output_file.associated_gene_list -post ankrd11filter -anno filtered.proband.annovar.hg19_multianno.txt
Then view the newly created file ankrd11filter and note the top 10 genes, and our causal gene ANKRD11 at number 1. It was previously number 2 if you check ankrd11/output_file.associated_gene_list.
Go to https://phen2gene.wglab.org. Click on the tab Patient notes:
and paste this paragraph of clinical notes in the text box:
The proband had an abnormally large fontanelle, which resolved without treatment. The proband does not appear to have a sacral dimple. Other than the presence of flat arches, there are no obvious signs of foot abnormalities. The proband does not look like his other siblings, although there was a resemblance between him and his sister when they were the same age. Features of proband’s face can be seen, including bushy eyebrows, broad nasal tip, short philtrum, full lips and cupid bow of upper lip. Video of proband’s behavior. Proband is non-verbal, and hyperactive. He repetitively spins his toy. While playing, he gets up from his chair, walks a few steps, stomps his feet, and sits back down. Additional interview in August 2015, after the parents received the proband’s diagnosis of KBG syndrome. The proband develops new mannerisms every four to six months, the most recent being short, hard breaths through the nose and head turning. The proband has had a substantial decrease in the number of seizures after starting an Epidiolex (cannabidiol) treatment (70-80% decrease as described by the parents). The frequency of seizures increased after the proband fell and fractured his jaw. The mother describes the proband’s macrodontia. Although the mother and several siblings have large teeth, the macrodontia in the proband does not appear in any other member of the family. The proband’s features are compared to other characteristics usually found in other KBG patients. Unlike most KBG patients, the proband has full lips. Like most KBG patients, the proband has curved pinkies (diagnosed as clinodactyly), which are often found in KBG patients. Although the proband has relatively short toes, this trait may have been inherited from the father. The proband also has curved toenails, which commonly appear in autistic children.
Then click Submit.
You should see that ANKRD11 is in the top 3. The reason it is not 2 as in the previous example is that we manually curated HPO terms from the patient notes for each patient in our Phen2Gene paper and this is raw notes with no curation done and all HPO terms are being automatically extracted by Doc2HPO. Despite this, the discrepancy in our results is minimal.
Alternatively, you can also submit the HPO terms from example/ANKRD11.txt manually using the tab HPO IDs (they are already the default terms in the website so you can just click Submit on that tab).
And you should see that ANKRD11 is still number 2.