OrthoFinder — Accurate inference of orthologs, orthogroups, the rooted species, gene trees and gene duplication events made easy!
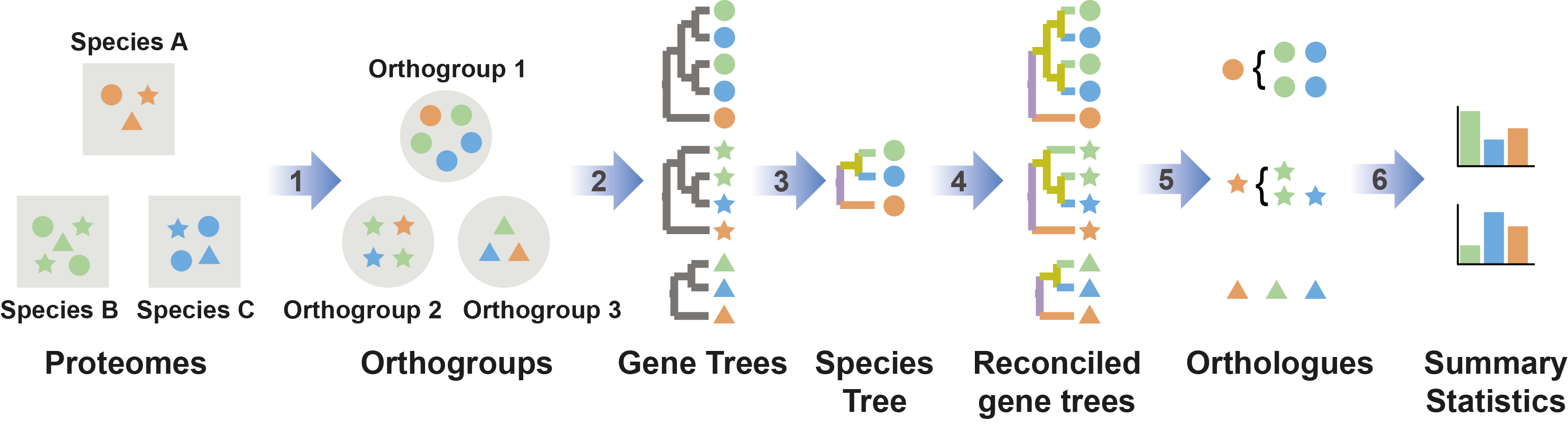 Figure 1: Automatic OrthoFinder analysis
Figure 1: Automatic OrthoFinder analysis
OrthoFinder is a fast, accurate and comprehensive platform for comparative genomics. It finds orthogroups and orthologs, infers rooted gene trees for all orthogroups and identifies all of the gene duplication events in those gene trees. It also infers a rooted species tree for the species being analysed and maps the gene duplication events from the gene trees to branches in the species tree. OrthoFinder also provides comprehensive statistics for comparative genomic analyses. OrthoFinder is simple to use and all you need to run it is a set of protein sequence files (one per species) in FASTA format.
For more details see the OrthoFinder papers below.
-
Download the latest release from github: https://github.com/davidemms/OrthoFinder/releases (for this example we will assume it is OrthoFinder-2.2.7.tar.gz, change this as appropriate.)
-
In a terminal, 'cd' to where you downloaded the package
-
Extract the files:
tar xzf OrthoFinder-2.2.7.tar.gz -
Install dependencies: MCL, FastME and DIAMOND (see below)
-
Test you can run OrthoFinder:
OrthoFinder-2.2.7/orthofinder -h. OrthoFinder should print its 'help' text.
To Run OrthoFinder on the Example Data type
OrthoFinder-2.2.7/orthofinder -f ExampleDataset -S diamond
A standard OrthoFinder run produces a set of files describing the orthogroups, orthologs, gene trees, resolve gene trees, the rooted species tree, gene duplication events and comparative genomic statistics for the set of species being analysed. These files are located in an intuitive directory structure.
-
Orthogroups.tsv is a tab separated text file. Each row contains the genes belonging to a single orthogroup. The genes from each orthogroup are organized into columns, one per species.
-
Orthogroups_UnassignedGenes.tsv is a tab separated text file that is identical in format to Orthogroups.csv but contains all of the genes that were not assigned to any orthogroup.
-
Orthogroups.txt (legacy format) is a second file containing the orthogroups described in the Orthogroups.tsv file but using the OrthoMCL output format.
-
Orthogroups.GeneCount.tsv is a tab separated text file that is identical in format to Orthogroups.csv but contains counts of the number of genes for each species in each orthogroup.
-
Orthogroups_SingleCopyOrthologues.txt is a list of orthogroups that contain exactly one gene per species i.e. they contain one-to-one orthologues. They are ideally suited to between-species comparisons and to species tree inference.
The Orthologues directory contains one sub-directory for each species that in turn contains a file for each pairwise species comparison, listing the orthologs between that species pair. Orthologues can be one-to-one, one-to-many or many-to-many depending on the gene duplication events since the orthologs diverged (see Section "Orthogroups, Orthologues & Paralogues" for more details). Each row in a file contains the gene(s) in one species that are orthologues of the gene(s) in the other species and each row is cross-referenced to the orthogroup that contains those genes.
- A phylogenetic tree inferred for each orthogroup
- A rooted phylogenetic tree inferred for each orthogroup and resolved using the OrthoFinder duplication-loss coalescent model.
-
SpeciesTree_rooted.txt A STAG species tree inferred from all orthogroups, containing STAG support values at internal nodes and rooted using STRIDE.
-
SpeciesTree_rooted_node_labels.csv The same tree as above but with the nodes given labels (instead of support values) to allow other results files to cross-reference branches/nodes in the species tree (e.g. location of gene duplication events).
-
Duplications_per_Orthogroup.tsv is a tab separated text file that gives the number of duplications identified in each orthogroup. This master file for this data is Gene_Duplication_Events/Duplications.tsv.
-
Duplications_per_Species_Tree_Node.tsv is a tab separated text file that gives the number of duplications identified as occurring along each branch of the species tree. This master file for this data is Gene_Duplication_Events/Duplications.tsv.
-
Orthogroups_SpeciesOverlaps.tsv is a tab separated text file that contains the number of orthogroups shared between each species-pair as a square matrix.
-
OrthologuesStats_*.tsv files are tab separated text files containing matrices giving the numbers of orthologues in one-to-one, one-to-many and many-to-many relationships between each pair of species.
-
OrthologuesStats_one-to-one.tsv is the number of one-to-one orthologues between each species pair.
-
OrthologuesStats_many-to-many.tsv contains the number of orthologues in a many-to-many relationship for each species pair (due to gene duplication events in both lineages post-speciation). Entry (i,j) is the number of genes in species i that are in a many-to-many orthology relationship with genes in species j.
-
OrthologuesStats_many-to-one.tsv: entry (i,j) gives the number of genes in species i that are in a one-to-many orthology relationship with genes from species j. There is a walk-through of an example results file here: davidemms#259.
-
OrthologuesStats_one-to-many.tsv: entry (i,j) gives the number of genes in species i that are in a many-to-one orthology relationship with a gene from species j. There is a walk-through of an example results file here: davidemms#259.
-
OrthologuesStats_Total.tsv contains the totals for each species pair of orthologues of whatever multiplicity. Entry (i,j) is the total number of genes in species i that have orthologues in species j.
-
-
Statistics_Overall.tsv is a tab separated text file that contains general statistics about orthogroup sizes and proportion of genes assigned to orthogroups.
-
Statistics_PerSpecies.tsv is a tab separated text file that contains the same information as the Statistics_Overall.csv file but for each individual species.
Most of the terms in the files 'Statistics_Overall.csv' and 'Statistics_PerSpecies.csv' are self-explanatory, the remainder are defined below.
- Species-specific orthogroup: An orthogroups that consist entirely of genes from one species.
- G50: The number of genes in the orthogroup such that 50% of genes are in orthogroups of that size or larger.
- O50: The smallest number of orthogroups such that 50% of genes are in orthogroups of that size or larger.
- Single-copy orthogroup: An orthogroup with exactly one gene (and no more) from each species. These orthogroups are ideal for inferring a species tree and many other analyses.
- Unassigned gene: A gene that has not been put into an orthogroup with any other genes.
-
Duplications.tsv is a tab separated text file that lists all the gene duplication events identified by examining each node of each orthogroup gene tree. The columns are "Orthogroup", "Species Tree node" (branch of the species tree on which the duplication took place, see Species_Tree/SpeciesTree_rooted_node_labels.txt), "Gene tree node" (node corresponding to the gene duplication event, see corresponding orthogroup tree in Resolved_Gene_Trees/); "Support" (proportion of expected species for which both copies of the duplicated gene are present); "Type" ("Terminal": duplication on a terminal branch of the species tree, "Non-Terminal": duplication on an internal branch of the species tree & therefore shared by more than one species, "Non-Terminal: STRIDE": Non-Terminal duplication that also passes the very stringent STRIDE checks for what the topology of the gene tree should be post-duplication); "Genes 1" (the list of genes descended from one of the copies of the duplicate gene), "Genes 2" (the list of genes descended from the other copy of the duplicate gene.
-
SpeciesTree_Gene_Duplications_0.5_Support.txt provides a summation of the above duplications over the branches of the species tree. It is a text file in newick format. The numbers after each node or species name are the number of gene duplication events with at least 50% support that occurred on the branch leading to the node/species. The branch lengths are the standard branch lengths, as give in Species_Tree/SpeciesTree_rooted.txt.
- A FASTA file for each orthogroup giving the amino acid sequences for each gene in the orthogroup.
- The same files as the "Orthogroup Sequences" directory but restricted to only those orthogroups which contain exactly one gene per species.
This contains all the files necessary for orthofinder to run. You can ignore this.
- What are orthogroups, orthologs & paralogs?
- Why use orthogroups in your analysis
- Installing Dependencies
- Adding and removing species from a completed OrthoFinder run
- Preparing and using separately run BLAST files
Orthologs are pairs of genes that descended from a single gene in the last common ancestor (LCA) of two species (Figure 2A & B). An orthogroup is the extension of the concept of orthology to groups of species. An orthogroup is the group of genes descended from a single gene in the LCA of a group of species (Figure 2A).
The example Figure 2 contains an orthogroup from three species, human, mouse and chicken. Human and mouse each have one gene in this orthogroup (HuA and MoA, respectively) while chicken has two genes (ChA1 and ChA2). The human and mouse genes are a pair of genes descended from a single gene in the last common ancestor of the two species, therefore these two genes are orthologs and there is a one-to-one orthology relationship between the two genes.
The two chicken genes arose from a gene duplication event after the lineage leading to chicken split from the lineage leading to human and mouse. As gene duplication events give rise to paralogs, ChA1 and ChA2 are paralogs of each other. However, both chicken genes are descended from a single gene in the last common ancestor of the three species. Therefore, both chicken genes are orthologs of the human gene and the mouse gene. Although they are orthologs, sometimes these complex relationships are referred to as co-orthologs (e.g. ChA1 and ChA2 are co-orthologs of HuA). In this case there is a many-to-one orthology relationship between the chicken genes and the human gene. There are only three kinds of orthology relationships one-to-one, many-to-one, and many-to-many. All of these relationships are identified by OrthoFinder.
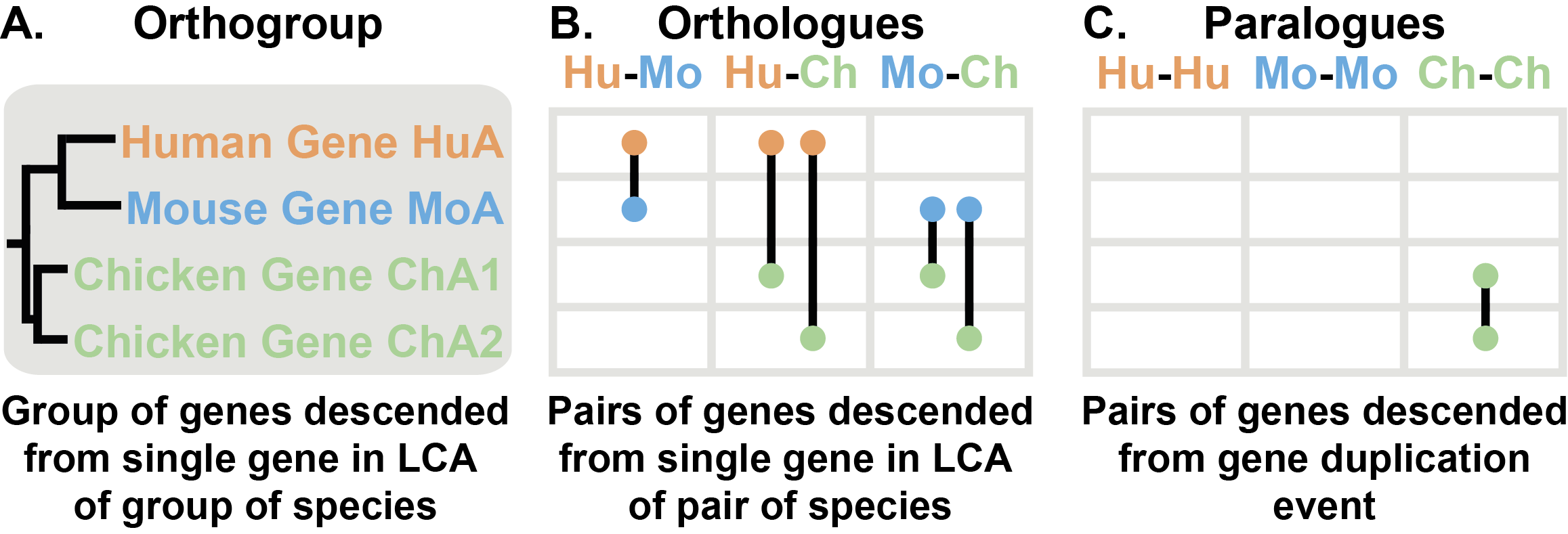 Figure 2: Orthologues, Orthogroups & Paralogues
Figure 2: Orthologues, Orthogroups & Paralogues
All of the genes in an orthogroup are descended from a single ancestral gene. Thus, all the genes in an orthogroup started out with the same sequence and function. As gene duplication and loss occur frequently in evolution, one-to-one orthologs are rare and limitation of analyses to on-to-one orthologs limits an analysis to a small fraction of the available data. By analysing orthogroups you can analyse all of your data.
It is important to note that with orthogroups you choose where to define the limits of the unit of comparison. For example, if you just chose to analyse human and mouse in the above figure then you would have two orthogroups.
Orthology is defined by phylogeny. It is not definable by amino acid content, codon bias, GC content or other measures of sequence similarity. Methods that use such scores to define orthologs in the absence of phylogeny can only provide guesses. To provide a crude analogy guessing orthology from sequence similarity is akin to guessing colour from smell. The only way to truly identify orthologs is thus through analysis of phylogenetic trees. The only way to be sure that the orthology assignment is correct is by conducting a phylogenetic reconstruction of all genes descended from a single gene the last common ancestor of the species under consideration. This set of genes is an orthogroup. Thus, the only way to define orthology is by analysing orthogroups.
To perform an analysis OrthoFinder requires some dependencies to be installed and in the system path (only the first two are needed to infer orthogroups and all four are needed to infer orthologues and gene trees as well):
-
DIAMOND or MMseqs2 (recommended, although BLAST+ can be used instead)
-
The MCL graph clustering algorithm
-
FastME (The appropriate version for your system, e.g. 'fastme-2.1.5-linux64', should be renamed `fastme', see instructions below.)
Brief instructions are given below although users can refer to the installation notes provided with these packages for more detailed instructions.
Available here: https://github.com/bbuchfink/diamond/releases
Download the latest release, extract it and copy the executable to a directory in your system path, e.g.:
wget https://github.com/bbuchfink/diamond/releases/download/v0.9.22/diamond-linux64.tar.gztar xzf diamond-linux64.tar.gzsudo cp diamond /usr/local/bin
or alternatively if you don't have root privileges, instead of the last step above, add the directory containing the directory to your PATH variable. E.g.
mkdir ~/bincp diamond ~/binexport PATH=$PATH:~/bin/
The mcl clustering algorithm is available in the repositories of some Linux distributions and so can be installed in the same way as any other package. For example, on Ubuntu, Debian, Linux Mint:
sudo apt-get install mcl
Alternatively, it can be built from source which will likely require the 'build-essential' or equivalent package on the Linux distribution being used. Instructions are provided on the MCL webpage, http://micans.org/mcl/.
FastME can be obtained from http://www.atgc-montpellier.fr/fastme/binaries.php. The package contains a 'binaries/' directory. Choose the appropriate one for your system and copy it to somewhere in the system path e.g. '/usr/local/bin'** and name it 'fastme'. I.e.:
sudo cp fastme-2.1.5-linux64 /usr/local/bin/fastme
NCBI BLAST+ is available in the repositories from most Linux distributions and so can be installed in the same way as any other package. For example, on Ubuntu, Debian, Linux Mint:
sudo apt-get install ncbi-blast+
Alternatively, instructions are provided for installing BLAST+ on Mac and various flavours of Linux on the "Standalone BLAST Setup for Unix" page of the BLAST+ Help manual currently at http://www.ncbi.nlm.nih.gov/books/NBK1762/. Follow the instructions under "Configuration" in the BLAST+ help manual to add BLAST+ to the PATH environment variable.
Available here: https://github.com/soedinglab/MMseqs2/releases
Download the appropriate version for your machine, extract it and copy the executable to a directory in your system path, e.g.:
wget https://github.com/soedinglab/MMseqs2/releases/download/3-be8f6/MMseqs2-Linux-AVX2.tar.gztar xzf MMseqs2-Linux-AVX2.tar.gzsudo cp mmseqs2/bin/mmseqs /usr/local/bin
or alternatively if you don't have root privileges, instead of the last step above, add the directory containing the directory to your PATH variable
export PATH=$PATH:`pwd`/mmseqs2/bin/
The following steps are not required for the standard OrthoFinder use cases and are only needed if you want to infer maximum likelihood trees from multiple sequence alignments (MSA). This is considerably more costly computationally but more accurate. By default, MAFFT is used for the alignment and FastTree for the tree inference. Both the executables should be in the system path. The option for this is, "-M msa".
You can actually use any alignment or tree inference program you like the best! Be careful with the method you chose, OrthoFinder typically needs to infer about 10,000-20,000 gene trees. If you have many species or if the tree/alignment method isn't super-fast then this can take a very long time! MAFFT + FastTree provides a reasonable compromise. Orthofinder already knows how to call:
- mafft
- muscle
- iqtree
- raxml
- raxml-ng
- fasttree
If you want to use a different program, there is a simple configuration file called "config.json" in the orthofinder directory. You just need to add an entry to tell it what the command line looks like for the program you want to use. There are lots of examples in the file that you can follow.
For example, to you muscle and iqtree, the command like arguments you need to add are: "-M msa -A muscle -T iqtree"
It is recommended that you use the standalone binaries for OrthoFinder which do not require python or scipy to be installed. However, the python source code version is available from the github 'releases' page (e.g. 'OrthoFinder-1.0.6_source.tar.gz' and requires python 2.7 and scipy to be installed. Up-to-date and clear instructions are provided here: http://www.scipy.org/install.html, be sure to choose a version using python 2.7. As websites can change, an alternative is to search online for "install scipy".
OrthoFinder allows you to add extra species without re-running the previously computed BLAST searches:
orthofinder -b previous_orthofinder_directory -f new_fasta_directory
This will add each species from the 'new_fasta_directory' to existing set of species, reuse all the previous BLAST results, perform only the new BLAST searches required for the new species and recalculate the orthogroups. The 'previous_orthofinder_directory' is the OrthoFinder 'WorkingDirectory/' containing the file 'SpeciesIDs.txt'.
OrthoFinder allows you to remove species from a previous analysis. In the 'WorkingDirectory/' from a previous analysis there is a file called 'SpeciesIDs.txt'. Comment out any species to be removed from the analysis using a '#' character and then run OrthoFinder using:
orthofinder -b previous_orthofinder_directory
where 'previous_orthofinder_directory' is the OrthoFinder 'WorkingDirectory/' containing the file 'SpeciesIDs.txt'.
The previous two options can be combined, comment out the species to be removed as described above and use the command:
orthofinder -b previous_orthofinder_directory -f new_fasta_directory
This functionality has been incorporated into the main 'orthofinder' program, replacing the old 'trees_from_MSA' utility. Trees can be inferred using MSAs by using the option "-M msa". If orthogroups have already been inferred then MSA trees can be inferred directly from them (rather than from inferring the orthogroups again from the start) by additionally using the option "-fg" option: "-M msa -fg previous_results_directory" instead of "-M msa -f input_proteomes_directory".
By default MAFFT is used to generate the multiple sequence alignments and FastTree to generate the gene trees. Alternatively, any other program can be used in place of these. Many popular programs have already been configured by having an entry in the config.json file in the orthofinder directory. All options currently available can be seen by using the option "-h" to see the help file. The config.json file is user-editable to allow for any other desired program to be added. MAFFT, FastTree, or whatever programs are used instead need to be in the system path.
There are two separate options for controlling the parallelisation of OrthoFinder. The '-t' option should always be used, typically with as many cores as are available. This determines how many highly-parallelisable tasks such as DIAMOND/BLAST searches, MSAs etc are run in parallel.
In addition, most of the internal steps of the OrthoFinder algorithm have been parallelised but by default run sequentially. Running these steps in parallel can be requested using the '-a' option. However, some of the steps can lead to large RAM requirements, potentially more than the computer has. This is the reason for this parallelisation being under separate control. These steps typically only make up a small part of the OrthoFinder runtime and so there is a lot less to gain from running these in parallel. If unsure, don't use this option.
-
'-t number_of_threads': This option should always be used. It makes the BLAST searches, the tree inference and gene-tree reconciliation run in parallel. These are all highly-parallelisable and the BLAST searches in particular are by far the most time-consuming task. You should use as many threads as there are cores available.
-
'-a number_of_orthofinder_threads' The remainder of the algorithm, beyond these highly-parallelisable tasks, is relatively fast and efficient and so this option has less overall effect. It is most useful when running OrthoFinder using pre-calculated BLAST results since the time savings will be more noticeable in this case. Using this option will also increase the RAM requirements (see manual for more details).
The '-op' option will prepare the files in the format required by OrthoFinder and print the set of BLAST commands that need to be run.
orthofinder -f fasta_files_directory -op
This is useful if you want to manage the BLAST searches yourself. For example, you may want to distribute them across multiple machines. Once the BLAST searches have been completed the orthogroups can be calculated using the '-b' command as described in Section "Using Pre-Computed BLAST Results".
It is possible to run OrthoFinder with pre-computed BLAST results provided they are in the correct format. They can be prepared in the correct format using the '-op' command and, equally, the files from a previous OrthoFinder run are also in the correct format to rerun using the '-b' option. The command is simply:
orthofinder -b directory_with_processed_fasta_and_blast_results
If you are running the BLAST searches yourself it is strongly recommended that you use the '-op' option to prepare the files first (see Section "Running BLAST Searches Separately"). Should you need to prepare them manually, the required files and their formats are described in the appendix of the PDF Manual (for example, if you already have BLAST search results from another source and it will take too much computing time to redo them).
A set of regression tests are included in the directory 'Tests' available from the github repository. They can be run by calling the script 'test_orthofinder.py'. They currently require version 2.2.28 of NCBI BLAST and the script will exit with an error message if this is not the case.