Wajid Jawaid 2021-02-02
enrichR can be installed from Github or from CRAN.
library(devtools)
install_github("wjawaid/enrichR")
The package can be downloaded from CRAN using:
install.packages("enrichR")
enrichR provides an interface to the Enrichr database [@kuleshov_enrichr:_2016] hosted at https://maayanlab.cloud/Enrichr/.
By default human genes are selected otherwise select your organism of choice. (This functionality was contributed by Alexander Blume)
library(enrichR)
listEnrichrSites()
#> Enrichr ... Connection is Live!
#> FlyEnrichr ... Connection is available!
#> WormEnrichr ... Connection is available!
#> YeastEnrichr ... Connection is available!
#> FishEnrichr ... Connection is available!
setEnrichrSite("Enrichr") # Human genes
#> Connection changed to https://maayanlab.cloud/Enrichr/
#> Connection is Live!
Then find the list of all available databases from Enrichr.
dbs <- listEnrichrDbs()
head(dbs)
| geneCoverage | genesPerTerm | libraryName | numTerms |
|---|---|---|---|
| 13362 | 275 | Genome_Browser_PWMs | 615 |
| 27884 | 1284 | TRANSFAC_and_JASPAR_PWMs | 326 |
| 6002 | 77 | Transcription_Factor_PPIs | 290 |
| 47172 | 1370 | ChEA_2013 | 353 |
| 47107 | 509 | Drug_Perturbations_from_GEO_2014 | 701 |
| 21493 | 3713 | ENCODE_TF_ChIP-seq_2014 | 498 |
View and select your favourite databases. Then query enrichr, in this case I have used genes associated with embryonic haematopoiesis.
dbs <- c("GO_Molecular_Function_2015", "GO_Cellular_Component_2015", "GO_Biological_Process_2015")
enriched <- enrichr(c("Runx1", "Gfi1", "Gfi1b", "Spi1", "Gata1", "Kdr"), dbs)
#> Uploading data to Enrichr... Done.
#> Querying GO_Molecular_Function_2015... Done.
#> Querying GO_Cellular_Component_2015... Done.
#> Querying GO_Biological_Process_2015... Done.
#> Parsing results... Done.
Now view the results table.
enriched[["GO_Biological_Process_2015"]]
You can give many genes.
data(genes790)
length(genes790)
head(enrichr(genes790, c('LINCS_L1000_Chem_Pert_up'))[[1]])
| Term | Overlap | P.value | Adjusted.P.value | Old.P.value | Old.Adjusted.P.value | Odds.Ratio | Combined.Score | Genes |
|---|---|---|---|---|---|---|---|---|
| embryonic hemopoiesis (GO_0035162) | 3/24 | 0.0e+00 | 0.0000083 | 0 | 0 | 951.0952 | 16465.833 | KDR;GATA1;RUNX1 |
| regulation of myeloid cell differentiation (GO_0045637) | 4/156 | 1.0e-07 | 0.0000083 | 0 | 0 | 261.0789 | 4374.968 | GFI1B;SPI1;GATA1;RUNX1 |
| regulation of erythrocyte differentiation (GO_0045646) | 3/36 | 1.0e-07 | 0.0000112 | 0 | 0 | 604.8788 | 9710.235 | GFI1B;SPI1;GATA1 |
| positive regulation of myeloid cell differentiation (GO_0045639) | 3/74 | 1.0e-06 | 0.0000762 | 0 | 0 | 280.6056 | 3886.803 | GFI1B;GATA1;RUNX1 |
| hemopoiesis (GO_0030097) | 3/95 | 2.1e-06 | 0.0001299 | 0 | 0 | 216.3261 | 2832.846 | KDR;GATA1;RUNX1 |
| hematopoietic progenitor cell differentiation (GO_0002244) | 3/106 | 2.9e-06 | 0.0001507 | 0 | 0 | 193.1165 | 2465.031 | SPI1;GATA1;RUNX1 |
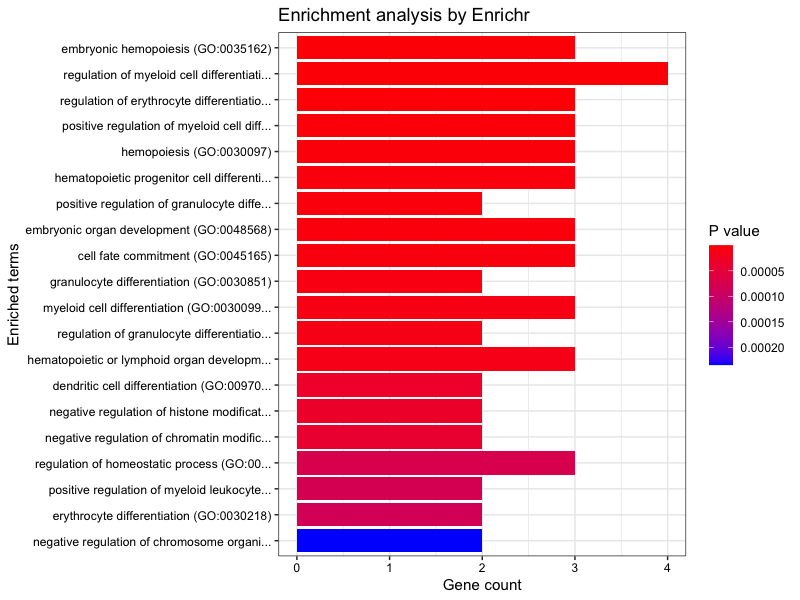
Plot Enrichr GO-BP output. (Plotting function contributed by I-Hsuan Lin)
plotEnrich(enriched[[3]], showTerms = 20, numChar = 40, y = "Count", orderBy = "P.value")
Kuleshov, Maxim V., Matthew R. Jones, Andrew D. Rouillard, Nicolas F. Fernandez, Qiaonan Duan, Zichen Wang, Simon Koplev, et al. 2016. “Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update.” Nucleic Acids Res 44 (Web Server issue): W90–W97. https://doi.org/10.1093/nar/gkw377.