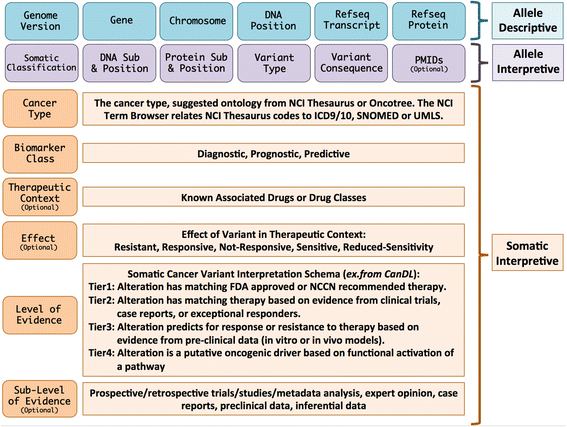
pyMVLD is a Python implementation of the Minimum Variant Level Data framework of standardized data elements, first defined by the ClinGen Somatic Working Group in 2016 (paper).
pyMVLD is an implementation of the MVLD framework for ensuring compliance. Use of this module provides:
- Validation of correct data types and values when generating MVLD objects
- Immutable, standardized objects for downstream applications
- Framework versioning
Creating an MVLD object first requires construction of three sub-objects corresponding to the three field sets described by the framework: AlleleDescriptive, AlleleInterpretive, and SomaticInterpretive.
The Allele Descriptive fields are expected to conform to the following rules:
The Genome Version must be in the GRCh37/GRCh38 format, preferably with the build version (e.g. GRCh38.p12). pyMVLD explicitly limits the values to a str describing GRCh37 or GRCh38, the two major GRC assemblies that are currently available. The assembly version, if provided, is checked only for syntactic correctness, not that it corresponds to a published version.
Genes must be provided as HGNC Approved Gene Symbols. The Gene field is checked to be a str, and is compared against the current HGNC list of approved symbols. If not HGNC Approved, the raised error describes the use of a known alias or retired symbol, if applicable.
kwargs = {
'genome_version': 'GRCh37',
'gene': 'BRAF',
'chromosome': 'chr7',
'dna_position': 'NC_000007.13:g.140453136A>T',
'refseq_transcript': 'NM_004333.4',
'refseq_protein': 'NP_004324.2'
}
ad = AlleleDescriptive(**kwargs)