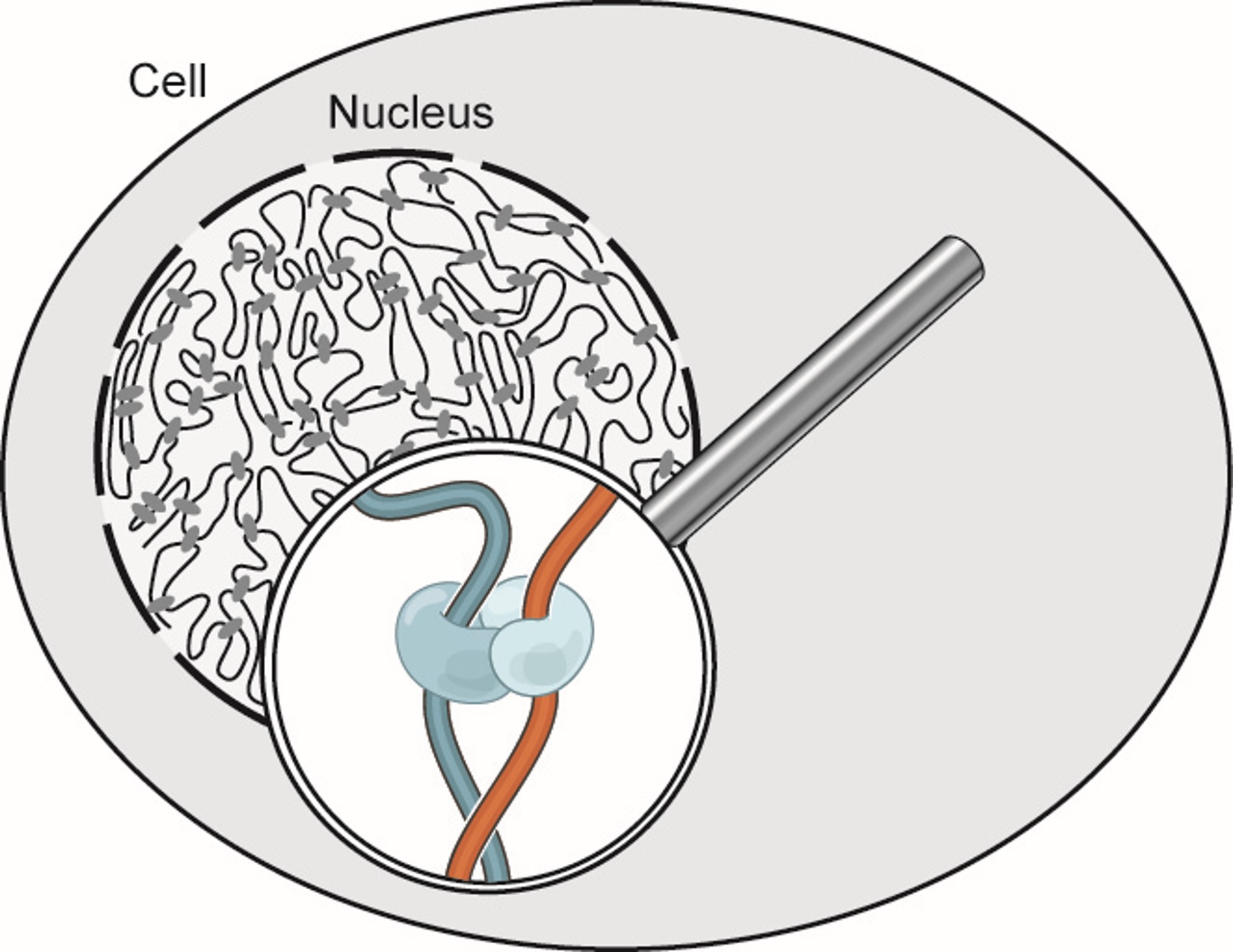
Every single cell in your body has the exact same DNA sequence, yet we observe vast differences in both structure and function within cells in our body. The differences between our transparent corneal cells and our highly conductive neurons exists not in the content of our DNA, but partly in how it is regulated.
This regulation is influenced by the 3D structure and organization of our chromatin, which is the uncoiled form of DNA present most of the cell's life cycle.
Therefore to understand the variations that exist between all life on earth, between the sick and healthy, and within ourselves, let us enter the microscopic world inside the nucleus and investigate the 3D organization of chromatin.
To study the 3D organization and structure of chromatin, we use Hi-C data.
Hi-C data represent one major approach for studying 3D interactions within the cell, using a technique known as contact mapping. Hi-C files are created by proximity ligation experiments (SEE MORE), which allows us to take "snapshots" of chromatin and cataloging what regions of the chromatin interact with each other. After a large amount of "snapshots", we can begin to get a complete picture of how the genome is organized.
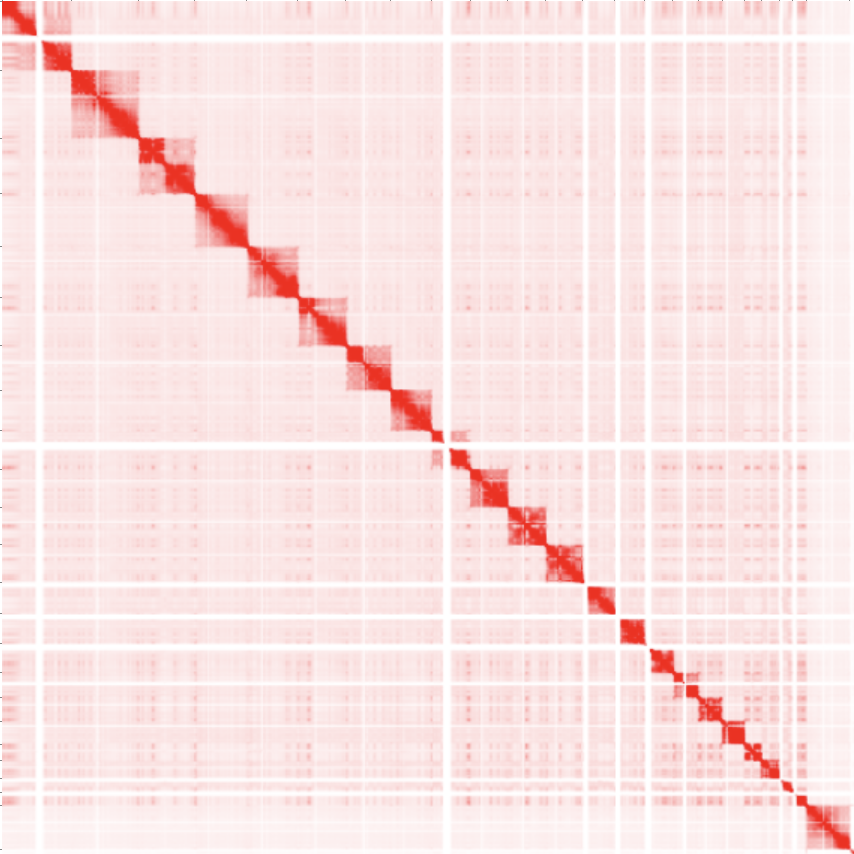
And this manifests itself as a Hi-C map! Below we can see an example of a typical genome wide Hi-C map. Notice how we can see distinct squares, those are the different chromosomes! The reason why they are more red (increased contacts) is because chromosomes tend to interact with themselves more than other chromosomes (chromosome territories)!
There are other genomic features other than chromosomes that can be visualized in Hi-C images like loops, domains and compartments. Keep reading to find out more!
For more information, See Dr. Erez Aiden's talk at the Scripps Research Translational Institute.
| Paper Name | Link |
|---|---|
| A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping | Link |
| Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes | Link |
| Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments | Link |
| Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom | Link |
| De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds | Link |
| Cohesin Loss Eliminates All Loop Domains | Link |