Project Overview:
This repository contains supplementary file, codebase and data generated for our paper titled "Mondrian Abstraction and Language Model Embeddings for Differential Pathway Analysis" which is currently under peer-review in a bioinformatics conference.
Supplementary File: The supplementary file to our paper can be found here.
Code: The notebooks folder contains the following jupyter notebooks:
- clinical_data_analysis.ipynb: Notebook for analyzing clinical data and suitable patient profile selection.
- data_preperation.ipynb: Notebook for preprocessing data to make it suitable for Mondrian Map Visualization.
- pathway_embeddings.ipynb: Here, we've experimented with different embedding techniques with different prompting strategies.
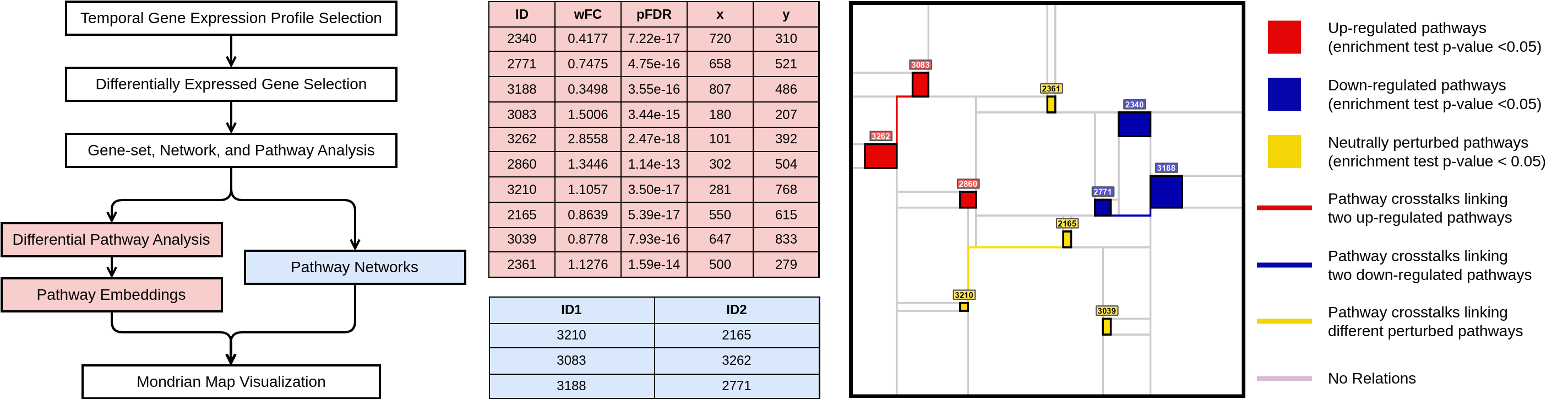
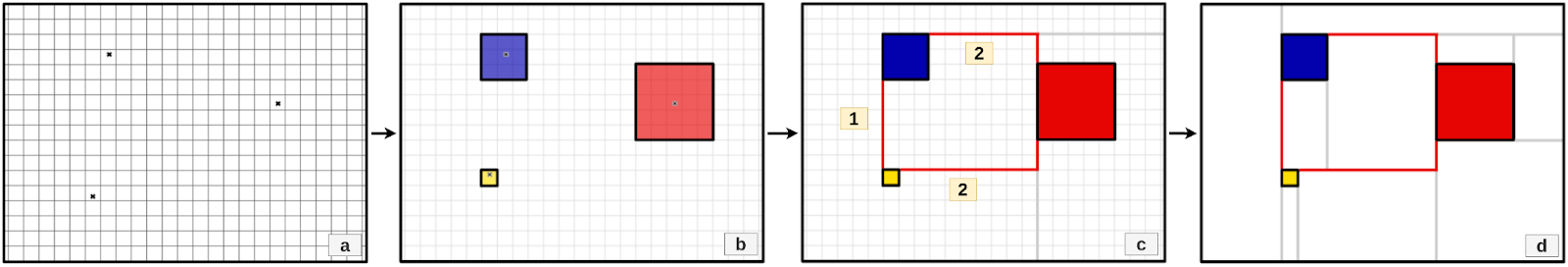
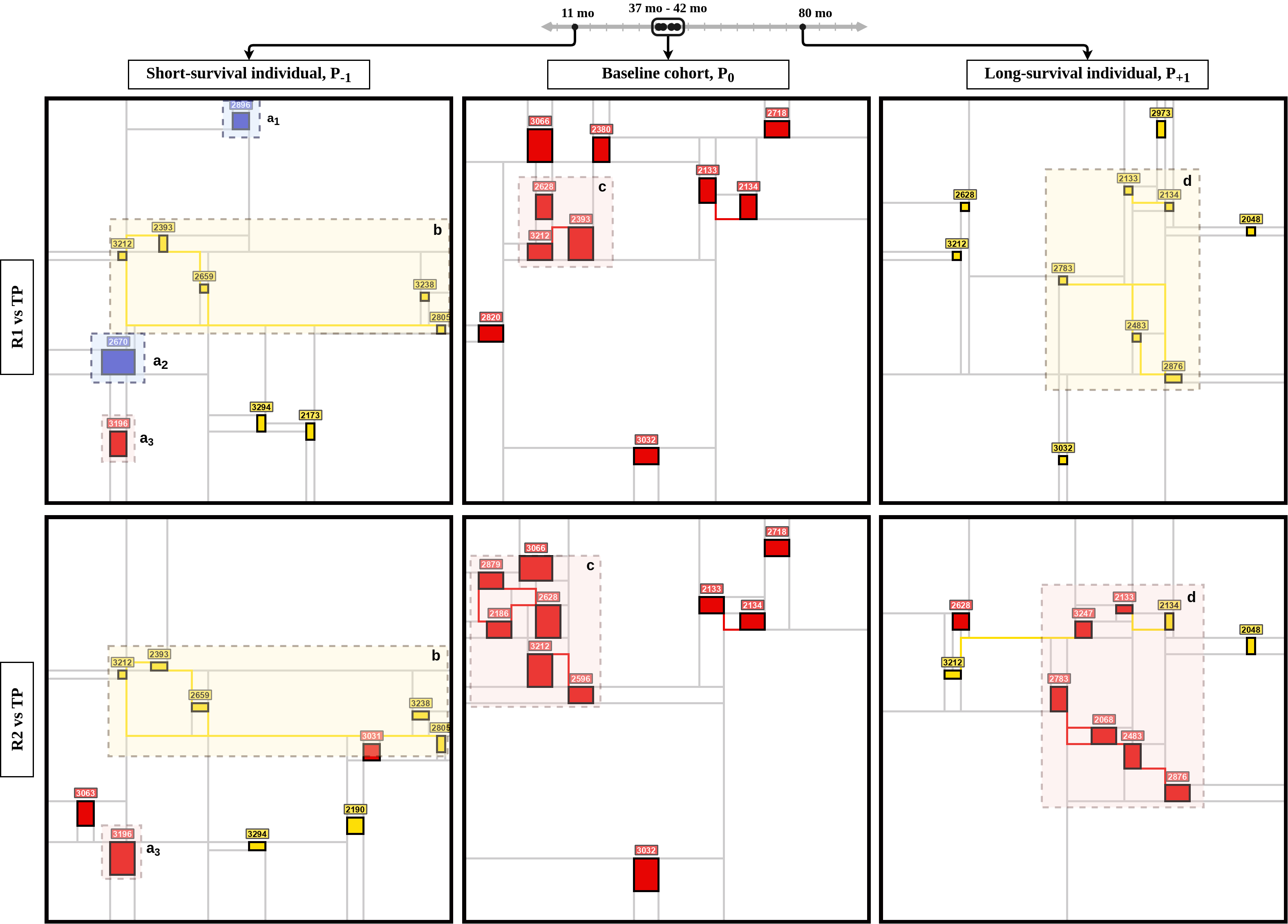
- visualize_mondrian_map.ipynb: In this notebook, we've generated the Mondrian Maps in our Gliblastoma case study.
Data: All the datasets used and processed are stored in the data folder.
If you find out tool useful, cite our latest preprint.
@article {AlAbir_MondrianMap,
author = {Al Abir, Fuad and Chen, Jake Y.},
title = {Mondrian Abstraction and Language Model Embeddings for Differential Pathway Analysis},
elocation-id = {2024.04.11.589093},
year = {2024},
doi = {10.1101/2024.04.11.589093},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/08/19/2024.04.11.589093},
eprint = {https://www.biorxiv.org/content/early/2024/08/19/2024.04.11.589093.full.pdf},
journal = {bioRxiv}
}
Reach us at jakechen@uab.edu or fuad021.edu.
Mondrian Map codebase is under MIT license.