cds_insertion is intended to add CDS features to transcriptome annotations lacking them (referred to later as the "raw" transcriptome annotation) (e.g. a GTF file generated by a transcript assembler such as stringtie (https://ccb.jhu.edu/software/stringtie/). cds_insertion accomplishes this by taking as input the raw transcriptome annotation as well as a separate transcriptome annotation that includes CDS features (e.g. GENCODE https://www.gencodegenes.org/), identifying overlaps between the raw transcriptome exons and annotated CDS start codons, and then translating the raw transcriptome transcripts (in silico) from the overlapping annotated start codons. Future versions will accept a bed file of start codon coordinates in lieu of a full annotation. The idea is to restrict inserted CDS features to those derived from start codons to those for which there is strong evidence of utilization (e.g. conservation, known protein sequences, evidence from Ribo-seq or similar) rather than inserting another layer of arbitrary decisions by performing de novo CDS predictions. cds_insertion additionally identifies a number of transcript properties derived from the presence/properties of the CDS such as the UTR lengths and the presence/absence of a premature termination codon.
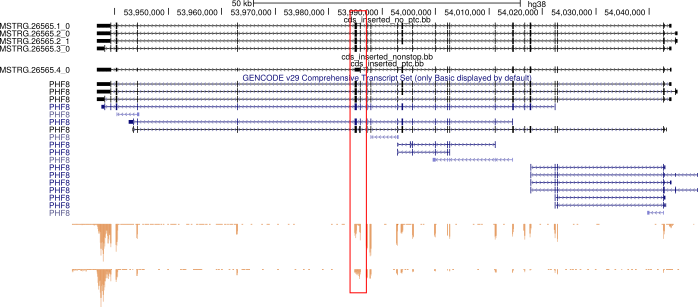
An example of an unannotated premature termination codon-containing transcript identified by running cds_insertion.py on a stringtie-augmented transcriptome GTF. The PTC is caused by an unannotated retained intron highlighted in red. Visualized bigGenePred file generated by cds_insertion (via Kent utils - see below) on the UCSC Genome Browser
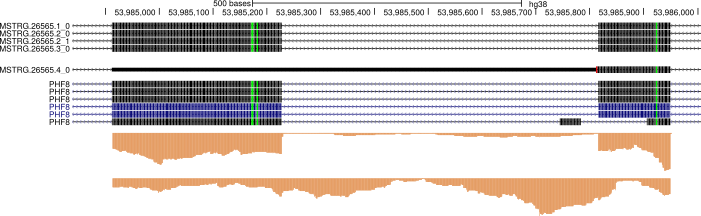
Zoomed-in view of the PTC-containing region described above
python 3
Bedtools v>=2.27
splice_lib (github.com/ajw2329/splice_lib/ - install using python setup.py install)
python /path/to/cds_insertion.py --transcript_gtf /path/to/raw/transcriptome.gtf --transcript_fasta /path/to/raw/transcriptome.fa --annotation_gtf /path/to/cds_annotated_transcriptome.gtf --outdir /path/to/output/`
Note that this assumes that bedtools is in your PATH. The location of the bedtools executable can be made explicit by adding --bedtools\_path /path/to/bedtools
The --CCDS flag will require that annotation CDS entries contain the text "CCDS". This is intended for use with a GENCODE annotation GTF, and will have the affect of using only start codons from consensus CDS features (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5753299/).
python /path/to/cds_insertion.py --transcript_gtf /path/to/raw/transcriptome.gtf --transcript_fasta /path/to/raw/transcriptome.fa --annotation_gtf /path/to/cds_annotated_transcriptome.gtf --outdir /path/to/output/ --CCDS`
This file provides basic transcript CDS information such as number of putative CDS, whether any truncated CDS are found (start codon with no in-frame stop), whether or not the transcript is a putative NMD substrate (based on max PTC distance >= 55nt), CDS lengths, UTR lengths, etc.
Here is an example of the file with field descriptions to follow:
| gene | transcript_id | normal_cds_count | nonstop_cds_count | always_nmd | sometimes_nmd | always_nonstop | sometimes_nonstop | PTC_distances | max_downstream_PTC_distances | CDS_lengths | five_utr_lengths | three_utr_lengths | three_utr_junction_counts | junction_contained_three_utr_lengths |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZNF746 | MSTRG.24752.1 | 1 | 0 | FALSE | FALSE | FALSE | FALSE | 0 | 0 | 1935 | 271 | 1591 | 0 | 0 |
| ZNF746 | MSTRG.24752.2 | 1 | 0 | FALSE | FALSE | FALSE | FALSE | 0 | 0 | 1980 | 271 | 1591 | 0 | 0 |
| ZNF746 | MSTRG.24752.3 | 1 | 0 | FALSE | FALSE | FALSE | FALSE | 0 | 0 | 1977 | 271 | 1591 | 0 | 0 |
| MSTRG.22509 | MSTRG.22509.1 | 0 | 0 | FALSE | FALSE | FALSE | FALSE | NA | NA | NA | NA | NA | NA | NA |
| MSTRG.28358 | MSTRG.28358.1 | 0 | 0 | FALSE | FALSE | FALSE | FALSE | NA | NA | NA | NA | NA | NA | NA |
| MSTRG.18752 | MSTRG.18752.1 | 0 | 0 | FALSE | FALSE | FALSE | FALSE | NA | NA | NA | NA | NA | NA | NA |
| MSTRG.11041 | MSTRG.11041.1 | 0 | 0 | FALSE | FALSE | FALSE | FALSE | NA | NA | NA | NA | NA | NA | NA |
| RGS5 | MSTRG.1910.1 | 2 | 0 | FALSE | FALSE | FALSE | FALSE | 0,0 | 0,0 | 219543 | 602278 | 5041,5041 | 0,0 | 0,0 |
| KRCC1 | MSTRG.14117.1 | 1 | 0 | FALSE | FALSE | FALSE | FALSE | 0 | 0 | 777 | 525 | 582 | 0 | 0 |
gene (string): gene symbol
transcript_id (string): transcript id
normal_cds_count (integer): number of "normal" (i.e. terminating in a stop codon rather than the end of the transcript) CDS found by translating overlapping annotated start codons
nonstop_cds_count (integer): number of "nonstop" (i.e. terminating in the end of the transcript rather than a stop codon) CDS found by translating overlapping annotated start codons
always_nmd (logical): TRUE/FALSE indicating whether the transcript contains a putative PTC with max distance >= 55 nt when translated from ALL overlapping annotated start codons
sometimes_nmd (logical): TRUE/FALSE indicating whether the transcript contains a putative PTC with max distance >= 55 nt when translated ANY overlapping annotated start codons (note that any transcript that is "always_nmd" is necessarily also "sometimes_nmd"
always_nonstop (logical): TRUE/FALSE indicating whether the ALL CDS generated by translating overlapping annotated start codons lack a stop codon (i.e. terminate at the 3'-end of the transcript)
sometimes_nonstop (logical): TRUE/FALSE indicating whether ANY CDS generated by translating overlapping annotated start codons lack a stop codon (note that "always_nonstop" transcripts are necessarily also "sometimes_nonstop")
PTC_distances (string containing comma-separated list of integers): distance of the putative PTC (if applicable) to the nearest downstream exon-exon junction. If there is more than one PTC (due to more than one CDS), all PTC distances are represented in a comma-separated list. Note also that non-PTC containing transcripts will have "0" in this field. In future iterations this may be changed to "NA".
max_downstream_PTC_distances (string containing comma-separated list of integers): distance of the putative PTC (if applicable) to the farthest downstream exon-exon junction. Note that these values are important because a PTC (for example) only 3 nt upsream of the nearest exon-exon junction may still satistfy NMD criteria if the farthest downstream exon-exon junction is >= 55 nt away. Even if the ribosome displaces the proximal EJC the remaining downstream EJC(s) may still aid in recruitment of NMD machinery. If there is more than one PTC (due to more than one CDS), all PTC distances are represented in a comma-separated list. Note also that non-PTC containing transcripts will have "0" in this field. In future iterations this may be changed to "NA".
CDS_lengths (string - comma separated list of integers): lengths of CDS found by translating overlapping annotated start codons
five_utr_lengths (string - comma separated list of integers): lengths of 5'-UTRs resulting from CDS found by translating overlapping annotated start codons
three_utr_lengths (string - comma separated list of integers): lengths of 3'-UTRs resulting from CDS found by translating overlapping annotated start codons
three_utr_junction_counts (string - comma separated list of integers): Number of exon-exon junctions in the 3'-UTR(s)
cds_inserted_no_ptc.gtf: GTF file of transcripts with inserted CDS features. Contains transcripts which do not have a putative PTC. Note that there is a separate set of entries (transcript, exon, CDS features) for each separate CDS associated with the transcript. These separate entries will be denoted by a number following the original transcript ID in the transcript_id feature. (e.g. if the transcript MSTRG.1.1 has two possible CDS they will be given separate entries MSTRG.1.1_0 and MSTRG.1.1_1)
cds_inserted_ptc.gtf: GTF file of transcripts with inserted CDS features. Contains transcripts with a putative PTC.
cds_inserted_nonstop.gtf: GTF file of transcripts with inserted CDS features. Contains transcripts with putative CDS that lack a stop codon (i.e. they terminate at the 3'-end of the transcript)
By default cds_insertion will output new GTF files with CDS entries, but for some visualization purposes it is nice to have bigGenePred files, which allow individual codon coloring when viewed on the UCSC genome browser. Generating the bigGenePred files requires the following UCSC utilities (by Jim Kent): bigGenePred, gtfToGenePred, bedToBigBed, and genePredToBigGenePred. These are available for linux and MacOS here: http://hgdownload.soe.ucsc.edu/admin/exe/. Provided those utilities are available in your PATH, the only necessary modifications to the above command are --make\_bigBed, --bigGenePred\_as_path /path/to/bigGenePred.as and --chrNameLength_path /path/to/chrNameLength.txt. The chrNameLength.txt file is just a two-column tsv containing chromosome name in the first column and chromosome length in the second. Note that this is just the first two columns of a .fai file such as is generated by samtools faidx. The bigGenePred.as file can be downloaded here: https://genome.ucsc.edu/goldenpath/help/bigGenePred.html.
The bigGenePred generation can be achieved (alongside the normal output) as follows:
python /path/to/cds_insertion.py --transcript_gtf /path/to/raw/transcriptome.gtf --transcript_fasta /path/to/raw/transcriptome.fa --annotation_gtf /path/to/cds_annotated_transcriptome.gtf --outdir /path/to/output/ --make_bigBed --bigGenePred_as_path /path/to/bigGenePred.as --chrNameLength_path /path/to/chrNameLength.txt`
Note that paths to the UCSC executables can be made explicit using --gtfToGenePred_path and equivalent for the rest.
usage: cds_insertion.py [-h] --transcript_gtf TRANSCRIPT_GTF
--transcript_fasta TRANSCRIPT_FASTA --annotation_gtf
ANNOTATION_GTF [--CCDS] --outdir OUTDIR
[--bigGenePred_as_path BIGGENEPRED_AS_PATH]
[--gtfToGenePred_path GTFTOGENEPRED_PATH]
[--genePredToBigGenePred_path GENEPREDTOBIGGENEPRED_PATH]
[--bedToBigBed_path BEDTOBIGBED_PATH]
[--bedtools_path BEDTOOLS_PATH]
[--chrNameLength_path CHRNAMELENGTH_PATH]
[--make_bigBed] [--ptc_dist PTC_DIST]
optional arguments:
-h, --help show this help message and exit
--transcript_gtf TRANSCRIPT_GTF
Input transcript gtf filename
--transcript_fasta TRANSCRIPT_FASTA
Input transcript fasta (assumes generation with
gffread)
--annotation_gtf ANNOTATION_GTF
Input gtf filename that contains CDS (preferably CCDS)
features
--CCDS If set, CDS used will be restricted to CCDS.
--outdir OUTDIR Path for output files
--bigGenePred_as_path BIGGENEPRED_AS_PATH
Path to bigGenePred.as file
--gtfToGenePred_path GTFTOGENEPRED_PATH
Path to gtfToGenePred exe
--genePredToBigGenePred_path GENEPREDTOBIGGENEPRED_PATH
Path to genePredToBigGenePred exe
--bedToBigBed_path BEDTOBIGBED_PATH
Path to bedToBigBed exe
--bedtools_path BEDTOOLS_PATH
Path to bedtools exe
--chrNameLength_path CHRNAMELENGTH_PATH
Path to chrNameLength file
--make_bigBed If set, will attempt to use Kent utils to output
bigbed file from gtfs. Requires chrNameLength_path and
bigGenePred_as_path to be set. If Kent utils are not
in PATH, set the paths to the executables as well.
--ptc_dist PTC_DIST Set minimum PTC distance required for transcript to be
considered putative NMD substrate. default = 55
find_switch_events is intended to identify alternative splicing events that switch between putative NMD or NSD substrates and non-substrates. The required input files are the .pkl file output by cds_insertion, an event gtf file (such as one generated by infer_pairwise_events.py), and an event ioe file (generated either by infer_pairwise_events.py or generateIOE.py). The script outputs a table with information on whether the event is an NMD or NSD switch event and whether (in the case of an NMD switch event) the alternative region of the putative NMD-substrate form contains the PTC.
python2.7
python find_switch_events.py --outdir /path/to/output_dir/ --ioe_file /path/to/splice_lib_events.ioe --event_gtf /path/to/splice_lib_events.gtf --transcript_dict_pkl /path/to/cds_insertion_transcript_dict.pkl
usage: find_switch_events.py [-h] [--suppress_output] [--outdir OUTDIR]
[--ioe_file IOE_FILE] [--event_gtf EVENT_GTF]
[--transcript_dict_pkl TRANSCRIPT_DICT_PKL]
optional arguments:
-h, --help show this help message and exit
--suppress_output If set, not file will be output
--outdir OUTDIR Output directory
--ioe_file IOE_FILE Event ioe file
--event_gtf EVENT_GTF
Event gtf file - required for certain features to work
e.g. PTC overlap determination if standard_event_dict
not supplied as argument to main()
--transcript_dict_pkl TRANSCRIPT_DICT_PKL
Pickled transcript dict from cds_insertion.py.
Required for certain features to work e.g. PTC overlap
determinatino if standard_transcript_dict not supplied
as argument to main()
generateIOE is a script that can be used to generate an event ioe file from an event gtf and a transcript gtf. While the ioe file is normally generated during creation of an event file, there are circumstances under which this cannot occur (such as when an event annotation is created by cross-species mapping of events).
python3
python generateIOE.py --transcript_gtf /path/to/transcriptome.gtf --event_gtf /path/to/splice_lib_events.gtf --outdir /path/to/output_dir/
| event_id | event_type | nmd_status | nmd_form | ptc_overlap | nonstop_status | nonstop_form |
|---|---|---|---|---|---|---|
| human_chimpanzee_orangutan_macaque|A5.0060093 | A5 | never | neither | NA | never | neither |
| human_chimpanzee_orangutan_macaque|RI.0044820 | RI | never | neither | NA | never | neither |
| human_chimpanzee_orangutan_macaque|SE.0023738 | SE | never | neither | NA | never | neither |
| human_chimpanzee_orangutan_macaque|A3.0022901 | A3 | always | excluded | FALSE | never | neither |
| human_chimpanzee_orangutan_macaque|RI.0077570 | RI | never | NA | NA | never | neither |
| human_chimpanzee_orangutan|RI.0036360 | RI | never | NA | NA | never | neither |
| human_chimpanzee_orangutan_macaque|SE.0023125 | SE | never | NA | NA | never | neither |
| human_chimpanzee_orangutan_macaque|SE.0059404 | SE | always | included | TRUE | never | neither |
event_id (string): unique identifier for each alt splicing event
event_type (categorical): Two-letter event-type identifier
nmd_status (categorical with possibilities always, sometimes, and never): Describes the putative NMD status of the event. always NMD events are those in which one isoform is only consistent with transcripts having a putative PTC (when translated from any overlapping annotated start codon provided in cds_insertion.py), while the other isoform is only consistent with events lacking a putative PTC. The behavior of these events is therefore expected to report on changes in NMD. sometimes NMD events fall into two categories. The first is similar to always NMD events, but the requirements are loosened: one of the isoforms must be consistent with at least one transcript with a putative PTC (again, translated using any putative ORF inserted by cds_insertion.py), while the other isoform must only be consistent with transcripts lacking a PTC. Consequently, these sometimes NMD events can still be assigned a particular nmd_form (included or excluded). The second category includes events in which at least one transcript from each isoform has a putative PTC. Consequently, nmd_form is given the value both, and there is no obvious way to predict the behavior of these events upon NMD perturbation. Note that the latter category of sometimes was previously identified as ambiguous_nmd. It was changed as the NMD status itself is not any more ambiguous than other sometimes events - it is instead the expected behavior of the event upon NMD perturbation whose behavior is ambiguous. Finally never NMD events are those in which no transcript consistent with either isoform is predicted to contain a PTC.
nmd_form (categorical with possibilities included, excluded, both, or neither): Indicates which event isoform is consistent with putative NMD substrates. This value is only supplied for events with always NMD or sometimes NMD status. Events with nmd_status always can only have nmd_form values included or excluded. Events with nmd_status sometimes can adopt all three nmd_form values. This variable is what allows prediction of event behavior under conditions of NMD perturbation. For example, we expect to observe a positive dPSI in events with nmd_form included upon NMD inhibition (e.g. through NMD factor knockdown or chemical inhibitors), and a negative dPSI under the same conditions for events with nmd_form excluded. Events with nmd_status never have nmd_form neither.
ptc_overlap (logical): Indicates whether the alternative region (e.g. the cassette exon in the case of a skipped exon (SE) event) contains all of the putative PTCs. It is only supplied for always events, and is otherwise NA.
nonstop_status (categorical with possibilities always, sometimes, and never): Indicates putative nonstop (referring to nonstop decay) switch status. always nonstop events are those in which one isoform is only consistent with transcripts whose complement of ORFs does not terminate in stop codons, while the other isoform is only consistent with transcripts whose complement of ORFs all contain stop codons. Similar to NMD events, sometimes events come in two flavors: 1) those in which one isoform is consistent with transcripts whose complement of ORFs sometimes lack a stop codon, while the other isoform is only consistent with transcripts lacking truncated ORFs, 2) those in which both isoforms are consistent with at least one transcript that contains at least one truncated ORF. The latter category of sometimes formerly had its own value ambiguous_nsd. never nonstop events are those in which neither isoform is consistent with any transcript that contains any truncated ORFs (given available data).
nonstop_form (categorical with possible values included, excluded, both, or neither): Similar to nmd_form. This indicates the form of always or sometimes events that is putatively an NSD substrate. both indicates both event isoforms are consistent with at least one transcript that is predicted to contain at least one truncated ORF. neither corresponds to all events with never nonstop status.
Much extended table that includes CDS lengths, CDS overlap, coding switch status etc. This is a superset of event_nmd_nsd_status.tsv. More details forthcoming.
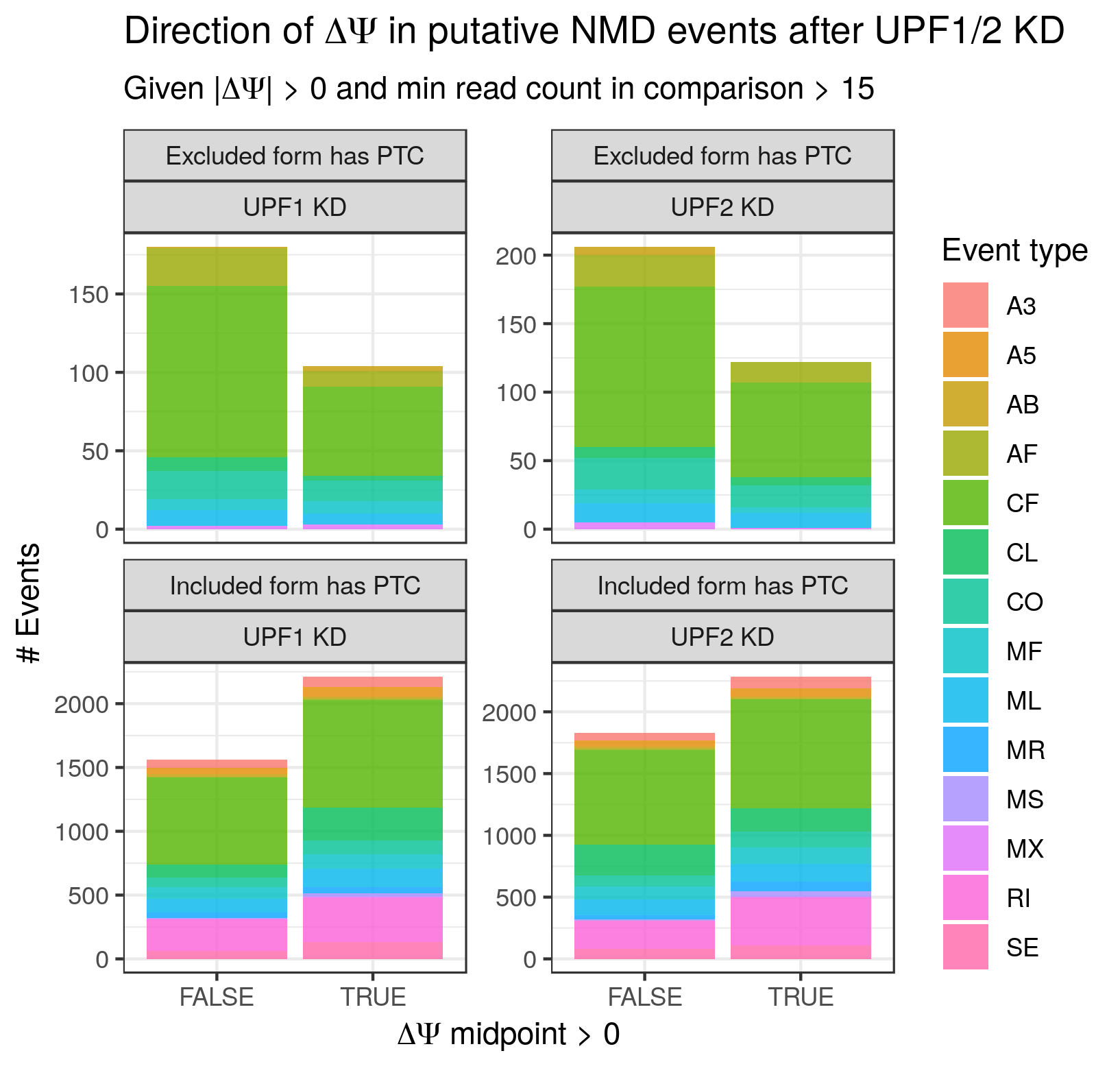
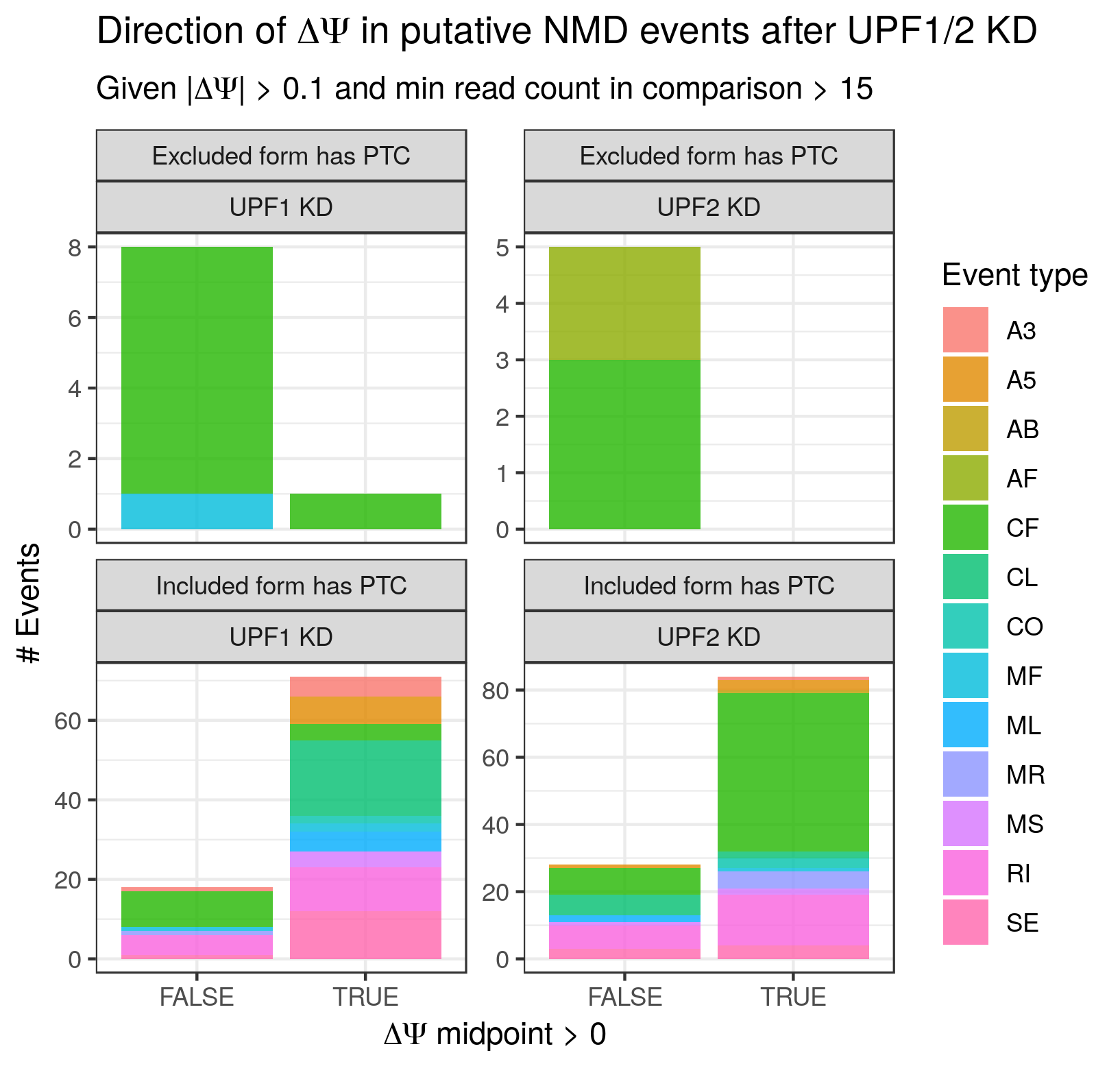
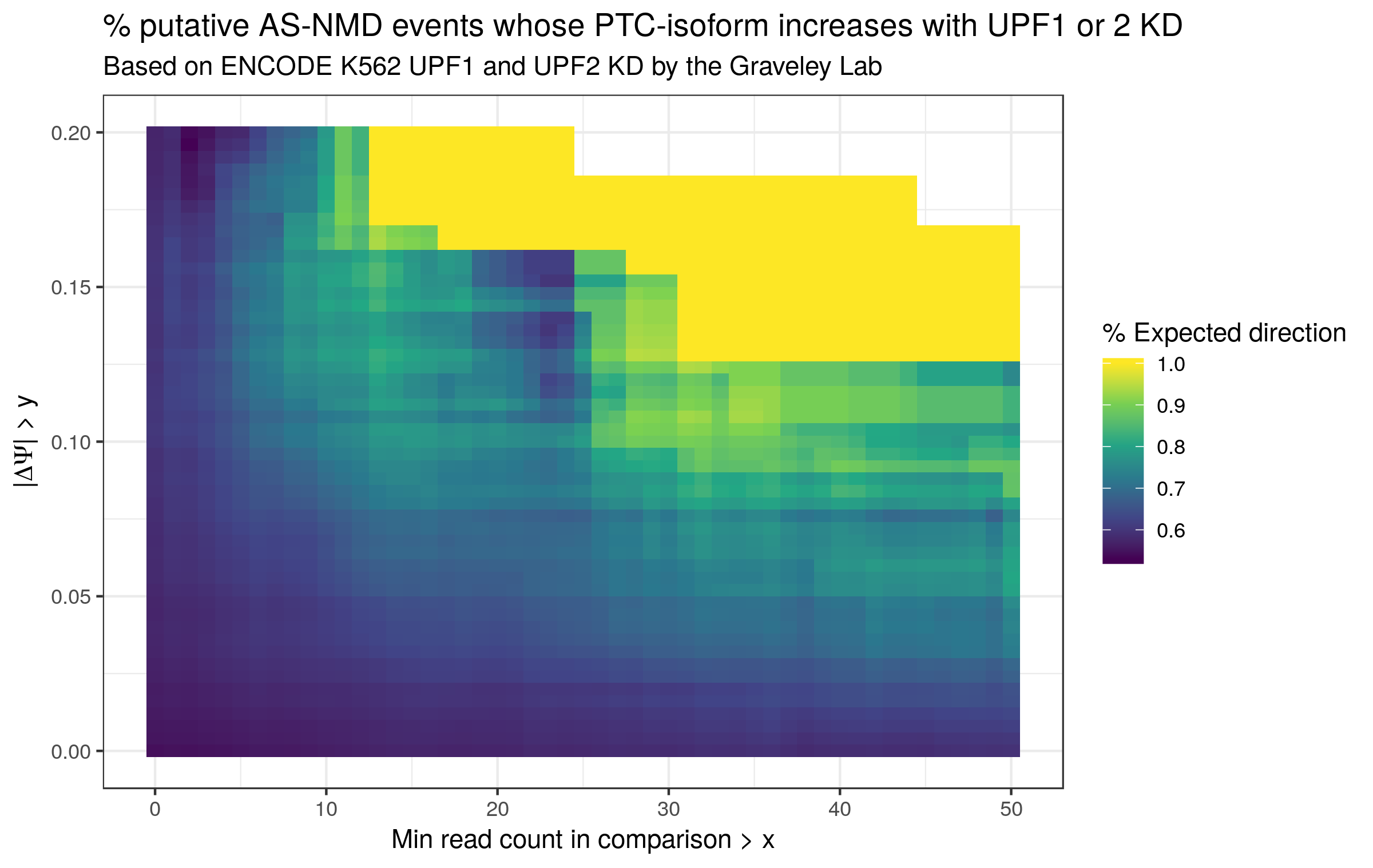
In order to ascertain whether find_switch_events.py is successful in identifying NMD switch events, I leveraged ENCODE RNA-seq data from the Gravely Lab derived from UPF1 and UPF2 KD K562 cells (found here and here for UPF1 and 2 respectively, with matched control data found here and here respectively). As UPF1 and 2 are part of the core mammalian NMD apparatus, the default expectation is that their knockdown will result in an increase in the abundance of PTC-containing transcripts. In the context of binary events such as those handled by find_switch_events.py, this translates to the expectation that NMD switch events whose included isoform contains (or is consistent with transcripts containing) PTC should exhibit a positive ΔΨ (change in percent-spliced-in) upon UPF1 or 2 KD, while those whose excluded isoform is the PTC-containing (or PTC-transcript-associated) isoform should exhibit a negative ΔΨ upon NMD factor KD. Granted, even to the extent that potential NMD switch events are successfully identified, their observed ΔΨ may not always match expectations for biological reasons. For example, they could be specifically protected from NMD in this particular context. Alternatively, the NMD isoform could become preferentially spliced out to compensate for its increased stability. Nevertheless, its my expectation that the observed ΔΨ values have the expected sign more often than not.
To test whether or not the ΔΨ values match the expectations described above, I mapped the data with STAR, ran stringtie (https://ccb.jhu.edu/software/stringtie/) to generate a transcriptome GTF (using GENCODE v29 Basic as a reference) and cds_insertion.py to annotated the new transcriptome with CDS features. I then ran junctionCounts (https://github.com/ajw2329/junctionCounts) to generate pairwise event annotations quantify the Ψ values for the events. Finally, I ran find_switch_events.py as well as custom R code (see the .Rmd file in ./validation) to annotate the NMD status of events and calculate ΔΨ values. I restricted the current analysis to the highest confidence class of putative NMD events - always_nmd events for which ptc_overlap == TRUE (i.e. the alternative region of the event overlaps the PTC). In future efforts I will explore the other classes.
The results are as follows: more often than not, the sign of the ΔΨ values (specifically the midpoint between the minimum and maximum possible ΔΨ for each event) matches the expected sign (given a non-zero ΔΨ). In Figure 1 below it can be seen that events whose included form contains a PTC and that exhibit a non-zero ΔΨ, the ΔΨ is positive more often than not, consistent with the assumption that the putative PTC-containing isoform's stability has increased. Similarly, for those events whose excluded form contains a PTC the ΔΨ is negative more often than not, consistent with the excluded isoform's PTC being less-efficiently subjected to NMD. The values in Figure 1 are restricted such that |ΔΨ| > 0, and the minimum number of junction reads found in any replicate in the comparison is 16. With these constraints, an aggregate of 58% of NMD switch events exhibit ΔΨ values that match expectations.
Figure 2 is essentially the same analysis, but with an increased |ΔΨ| threshold (> 0.1). With this new threshold, the percentage of NMD switch events whose ΔΨ sign matches expectation increases to 78%. Figure 3 is a heatmap of the percentage calculation as a function of both |ΔΨ| and junction count thresholds, and it is clear that as these thresholds increase, so too does the percentage that matches expectations (with the caveat that the number of events being examined is decreasing). It is broadly my expectation that the minimum observed junction counts and the minimum |ΔΨ| speak to the reliability of the quantification and the magnitude of the change. The increase in percentage with increasing thresholds suggests to me that the more trustworthy the quantifications the better they supported by the find_switch_event.py annotations.
usage: generateIOE.py [-h] [--transcript_gtf TRANSCRIPT_GTF]o_access_the_public_servers
[--event_gtf EVENT_GTF] [--outdir OUTDIR]
[--prefix PREFIX]/people/anjowall/projects/primate_neurodiff/
optional arguments:
-h, --help show this help message and exit
--transcript_gtf TRANSCRIPT_GTF
Full transcript gtf file
--event_gtf EVENT_GTF
Event gtf
--outdir OUTDIR Path to output directory
--prefix PREFIX Prefix for output files