OptiScan is an open source tool for extraction of nick and backbone signals from provided Bionano Genomics optical map TIFF images. OptiScan is able to transform nick signals into nicking site coordinates and output these in the BNX format for further processing.
Python >=3.6sqlite
git clone https://gitlab.com/akdel/OptiScan.git
cd OptiScan
pip install .OptiScan dashboard is only available for Irys platform. See command-line version for Saphyr data.
- Run the http server with permitting external access (or use
--ip=localhostfor local access) and a desired port:optiscan_dashboard --ip=0.0.0.0 --port=8080
- Access it from a browser at
http://0.0.0.0:8080/optiscan(orhttp://localhost:8080/optiscan).
While ordinarily OptiScan is best used from the command line, a dashboard is also available.
It is built as a web application with
Dash. In this interface, you can
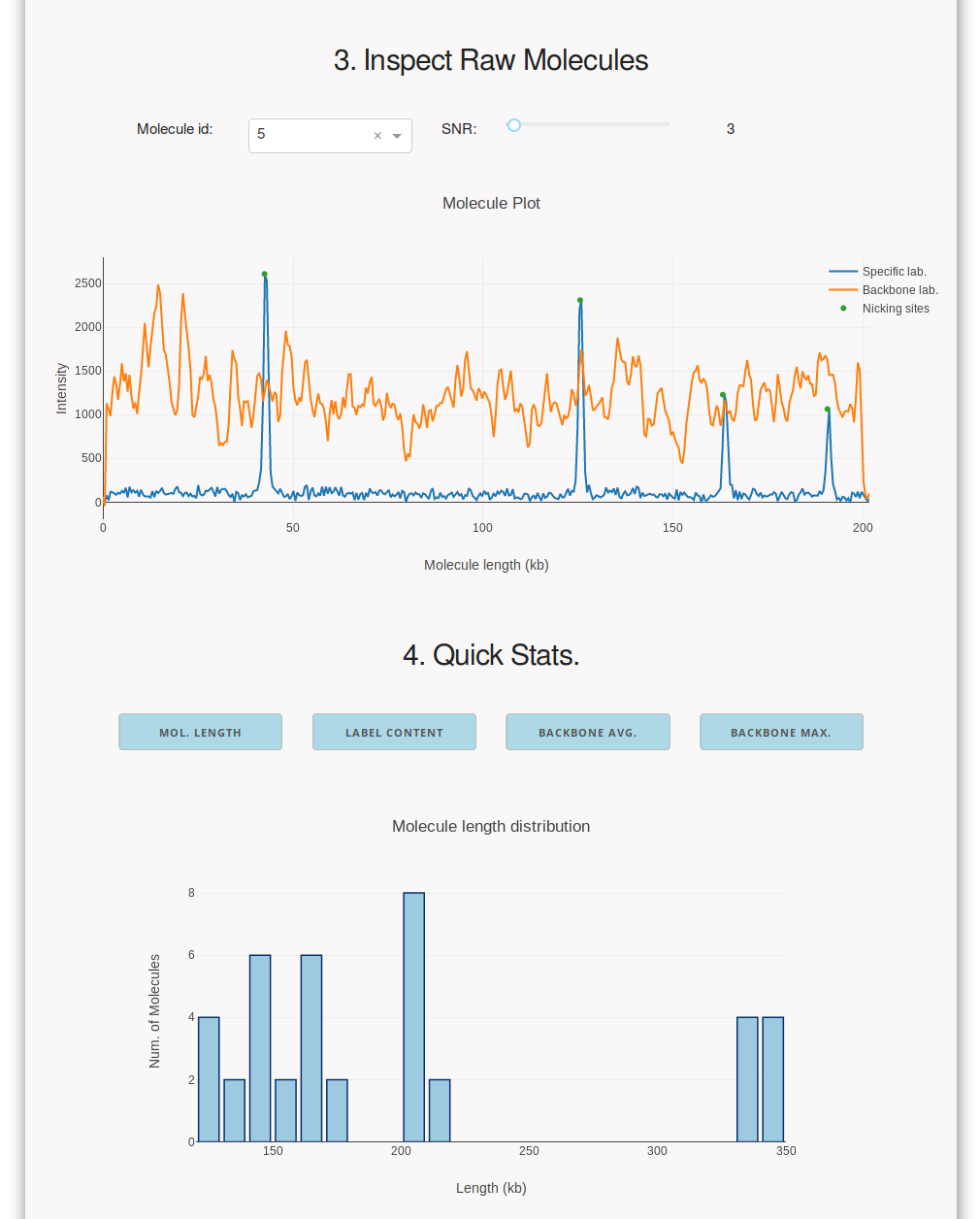
execute molecule detection/extraction and inspect the raw molecules and
molecule length distributions. Inspecting these values supports the choice
of SNR, maximum DNA backbone intensity and minimum molecule length
thresholds prior to exporting the data in the BNX format.
You can find two scan image files in the directory data/test_run/ and specify this
path in the dashboard under Run Folders.
Providing all the test parameters in Molecule detection section and running OptiScan is required to extract molecules and create the test database. Test parameters are already filled (except the database name) when the dashboard is launched. Molecules in the test data can be simply detected by entering a database name and clicking RUN OPTISCAN. This process can take up to 5-10 minutes to complete, depending on your workstation speed.
You can find the live demonstration of OptiScan dashboard at http://bioinformatics.nl/optiscan/. (note: the demo interface lacks the molecule detection function.)
The optiscan/pipelines/ folder contains several scripts for running OptiScan with Irys or Saphyr data.
-
extract_molecules_irysandextract_molecules_saphyrare for extracting and recording optical map signals. (Details on parameters and usage are given with cli--help) example usage:extract_molecules_irys '/path/to/run1,/path/to/run/2' '12,95' --database_name=data.db --number_of_threads=10 extract_molecules_saphyr '/path/to/saphyr_data' --number_of_threads=10
Usage with test data: Test data scans are reduced in dimension to 12x1 frames. To run the test data, simply provide
"12,1"as--chip_dimension:extract_molecules_irys 'data/test_run' '12,1' --database_name=test.db --number_of_threads=2
-
write_bnx_irysandwrite_bnx_saphyrare for writing extracted molecules in the BNX format. (Details on parameters and usage are given with cli--help)example usage:
write_bnx_irys /path/to/apple.db --snr=3 write_bnx_saphyr /path/to/saphyr_data --snr=3
extract_molecules_irys 'data/test_run' '12,1' --database_name=test.db --number_of_threads=2