The scribl language was designed for the curation from scientific articles, of the relationships between the various biological agents and processes that they describe, with a view to generating a graph database that captures these relationships as a connected network.
scribl statements are added as tags to articles in a literature database (the scribl codebase currently supports the free, open-source Zotero database). These tags are parsed and used to create a graph data structure that can be then be exported for use in a graph database platform such as neo4j, or Python's NetworkX.
For example:
::agent c9orf72 :gene :protein :url https://www.uniprot.org/uniprot/Q96LT7
::agent gtp :tag nucleoside, purine, nucleoside triphosphate
::process exportin releases cargo into cytoplasm @ exportin-1
::process smcr8 mutation > ulk1 phosphorylation < autophagy = smcr8 expression
Full details of the language are available in the full documentation contained in scribl.pdf.
If you write a paper that uses scribl in your analysis, please cite
both:
-
our 2024 article published in the Journal of Open Source Software:
Webster GD, Lancaster AK. (2024) "scribl: A system for the semantic capture of relationships in biological literature." Journal of Open Source Software 9(99):6645. doi:10.21105/joss.06645
-
and the Zenodo record for the software. To cite the correct version, follow these steps:
-
First visit the DOI for the overall Zenodo record: 10.5281/zenodo.12728362. This DOI represents all versions, and will always resolve to the latest one.
-
When you are viewing the record, look for the Versions box in the right-sidebar. Here are listed all versions (including older versions).
-
Select and click the version-specific DOI that matches the specific version of scribl that you used for your analysis.
-
Once you are visiting the Zenodo record for the specific version, under the Citation box in the right-sidebar, select the citation format you wish to use and click to copy the citation. It will contain link to the version-specific DOI, and be of the form:
Webster GD, Lancaster, AK. (YYYY) "scribl: A system for the semantic capture of relationships in biological literature" (Version X.Y.Z) [Computer software]. Zenodo. https://doi.org/10.5281/zenodo.XXXXX
Note that citation metadata for the current Zenodo record is also stored in CITATION.cff
-
We recommend installing in a virtual environment.
pip install scriblgit clone https://github.com/amberbiology/scribl.git
cd scribl
pip install .This will pull in all relevant Python dependencies, including pyparsing, pyzotero and others within your virtual environment
When scribl is installed, it contains a command-line program scribl
that can perform some common API tasks, and to help the user jumpstart
exploring using the system. After installation here are two example
command-line you can cut-and-paste and run from the terminal to check
installation:
scribl --versionFirst create a new, empty directory and change into it, e.g.:
mkdir /tmp/scribl-test
cd /tmp/scribl-testThen run:
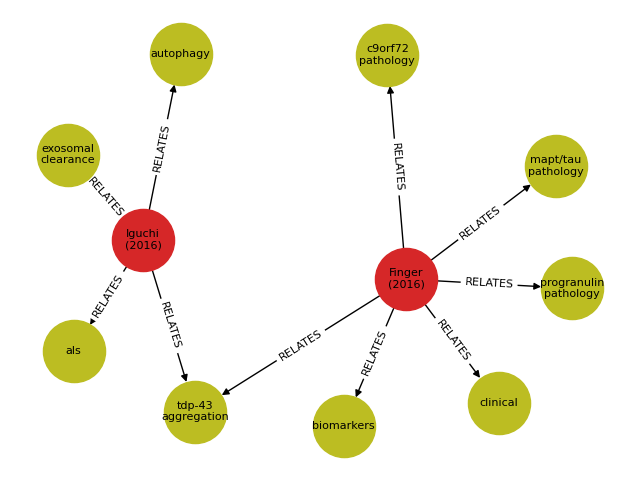
scribl -g new_graphdb --zotero-library 5251557:group --networkx-fig graphdb-visual.png --graphmlfile graphdb.xml --cyphertextfile graphdb.cypherThis will read from a public Zotero group collection we have created
(ID 5251557) for testing purposes, that includes citations with
scribl tags. This example run generates the following outputs:
-
new_graphdb- directory containing the new scribl database -
graphdb.cypher- a plain text file in the Cypher query language -
graphdb.xml- an XML file in GraphML format -
graphdb-visual.png- a visualization of the GraphML graph in PNG format:see below for a more detailed description of visualization options.
As mentioned previously, a scribl database can be exported for use
in a graph database platform for further interactive exploration. One
of the most developed graph visualization platforms is as
Neo4j which reads the output in Cypher query
language . Unfortunately Neo4j isn't
directly available via Python packaging system. The instructions for
installation and configuration is beyond the scope of this
documentation, but they are available for most
platforms.
The main scribl documentation describes how to take the
exported Cypher text generated by a command like the above into Neo4j
and dynamically update the Neo4j database.
In the interests getting up and running quickly with visualizations,
but staying purely within the Python ecosystem, scribl also supports
outputs other than Cypher. The above mentioned
GraphML backend generates
visualizations using a combination of
NetworkX and matplotlib. This feature enables
generation of static visualizations of the current scribl database.
Visualizations generated via the GraphML/NetworkX are best suited for
small networks and to get a feel for the scribl database structure
with a minimum of fuss. For "production" level visualization as well
as interactive analyses, however, you are likely to want to try Neo4j.
Here are some common scribl tasks you can do using the scribl
command-line:
scribl -g new_graphdb --zotero-library <LIBRARY_ID>:<TYPE> --zotero-api-key <API_KEY>A scribl database is created in current directory with the name
new_graphdb using the Zotero LIBRARY_ID with specified TYPE
(either user or group). An API_KEY is only needed in the case of
accessing private libraries, or non-public group libraries. The
pyzotero library has
documentation on
how to find out your library ID and creating an API key.
scribl -g new_graphdb --zoterofile <ZOTERO_CSV>This populates the database from a local CSV file ZOTERO_CSV exported
from a Zotero library. Note that the above two commands, just create (or
read from an existing) database, but generate no output. (Note that the
new_graphdb database is not overwritten if it already exists, unless the
--overwrite flag is also supplied)
To generate outputs, there are a number of options. All assume at least one Zotero database has been imported, either from a local CSV file, or via the Zotero library API (i.e. at least one of the import steps above have been run at least once). Outputs will be skipped if the database empty.
scribl -g new_graphdb --graphmlfile <OUTPUT_XML>This generates output in GraphML XML format (e.g. OUTPUT_XML should
be supplied with the appropriate extension, e.g. graphdb.xml).
scribl -g new_graphdb --networkx-fig <OUTPUT_IMAGE>This generates and saves the visualization as as one of the
matplotlib supported OUTPUT_IMAGE formts, using data from current
scribl database (for example, using graphdb-visual.pdf would
generate it in PDF format). The visualizations are produced from
GraphML XML output that is rendered using Python's NetworkX
library.
scribl -g new_graphdb --cyphertextfile <OUTPUT_CYPHER>The above outputs a Cypher text representation of the database in the
file OUTPUT_CYPHER, as a plain text file in the previously mentioned
Cypher query language, suitable for import
into Neo4J.
For a full description of all command-line arguments, run:
scribl --helpYou can write your own Python program using the API directly. For
example, the code below, generates the same outputs as the check
outputs example above. It creates a new database,
populates it from the same remote Zotero library, and then generates
graphdb.xml (GraphML), graphdb.cypher (Cypher) and
graphdb-visual.png (PNG).
from scribl.manage_graphdb import GraphDBInstance
gdb = GraphDBInstance("newgraph_db")
gdb.set_metadata("Test Scribl DB", "User name", "Small test")
gdb.import_zotero_library(5251557, "group")
gdb.load_zotero_csv()
gdb.export_graphml_text(filepath="graphdb.xml")
gdb.export_graphml_figure(filepath="graphdb-visual.png")
gdb.export_cypher_text(filepath="graphdb.cypher")The code for the scribl command contains more
examples of use of the API. In addition, more details can be found
in the main documentation.
-
clone the repo:
git clone https://github.com/amberbiology/scribl.git
-
install with
pipwith the optiontestpackage :pip install .[test]
-
run
pytestNote that
pytestcreates output sandbox test files in the system temporary directory, e.g. on Linuxpytestcreates a directory structure/tmp/pytest-of-USER/pytest-NUM/scribl_sandboxNUMwhereUSERis the current user andNUMis incremented on each run. Only the last three directories are retained.
scribl is completely open-source and being developed by Amber Biology LLC (@amberbiology).
See CHANGELOG.md for a history of changes.
If you're interested in contributing, please read our CONTRIBUTING guide.
scribl is Copyright (C) 2023, 2024. Amber Biology LLC
scribl is distributed under the terms of AGPL-3.0 license
The development of the scribl platform was made possible by the funding and expertise provided by the Association for Frontotemporal Degeneration (AFTD - https://www.theaftd.org/) whose mission is to improve the quality of life of people affected by Frontotemporal Degeneration and drive research to a cure. We are particularly grateful to AFTD leadership team members Debra Niehoff and Penny Dacks for their invaluable direction and guidance, and for getting us access to leading researchers in the field of neurodegenerative disease - all of whose insights and advice were pivotal in the development of this free, open-source research tool.