This repository supports our recent MedNeurIPS 2022 abstract on the importance of context versus foreground-to-background ratio in segmentation tasks. See here for the abstract.
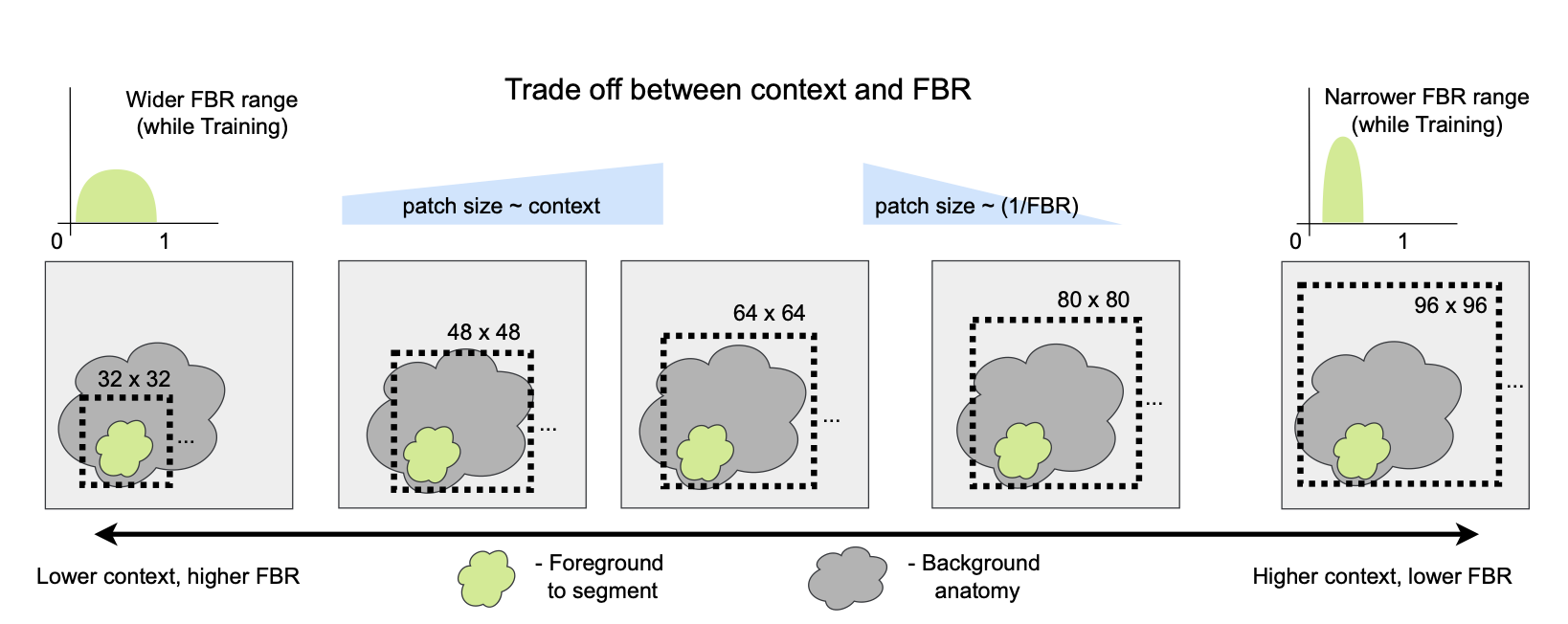
Modern 3D medical image segmentation is typically done using a sliding window approach due to GPU memory constraints. However, this presents an interesting trade-off between the amount of global context the network sees at once, versus the proportion of foreground voxels available in each training sample. It is known already that Unets perform worse with low global context, but enlarging the context comes at the cost of heavy class imbalance between background (typically very large) and foreground (much smaller) while training. In this abstract, we analyze the behavior of Transformer-based (UNETR) and attention gated (Attention-Unet) models along with vanilla-Unets across this trade-off. We explore this using a synthetic data set, and a subset of the spleen segmentation data set from the Medical Segmentation Decathlon to demonstrate our results. Beyond showing that all three types of networks prefer more global context rather than bigger foreground-to- background ratios, we find that UNETR and attention-Unet appear to be less robust than vanilla-Unet to drifts between training versus test foreground ratios.
To reproduce these results, run the following:
run_synthetic_experiments.sh
and
run_clinical_experiments.sh
The dependencies for this project are in the requirements file. They are listed below as well:
monai==0.9.1
nibabel==4.0.2
numpy==1.23.2
torch==1.12.1
wandb==0.13.3
The clinical data comes from the Medical Segmentation Decathlon, specifically the spleen data set. This data can be downloaded from the organizers' google drive and needs to be placed in this structure:
context_vs_fbr
│ README.md
│ ...
└───data
│ └─── raw
| └─── Task09_Spleen
└───spleen_experiments
| │ spleen_3d_wandb_attention_unet.py
| | ... unetr.py
| └─── ... unet.py
└───synthetic_experiments
| │ synthetic_3d_wandb_attention_unet.py
| | ... unetr.py
| └─── ... unet.py
...
The synthetic data will be generated automatically by the scripts in the synthetic_experiments folder and will also be uploaded to wandb for archiving.
Run the shell scripts as indicated earlier, and then to analyze the results, you could download the CSV files from the individual runs of the test prediction tables on wandb, for example: here - look under tables for the 'Export as CSV' option.
Then run the notebooks to generate the graphs, for synthetic experiments:
synthetic-analysis.ipynb
and for clinical (spleen) experiments:
spleen-analysis.ipynb
We use weights and biases to log and track the results of all our experiments. The following public links point to the runs we use for making inferences in this abstract, from where the CSV files are extracted to produce the results and figures used in the paper. These CSV files are copied over in this repository for easy access, but should be downloadable from these runs independently as well.
Attention-Unet: wandb project
UNETR: wandb project
Vanilla-Unet: wandb project
Attention-Unet: wandb project
UNETR: wandb project
Vanilla-Unet: wandb project
Please create an issue in this repository, or contact Amith here.
We gratefully acknowledge MONAI and wandb for all the great work they've done to create reusable libraries for network construction and reproducible analysis.