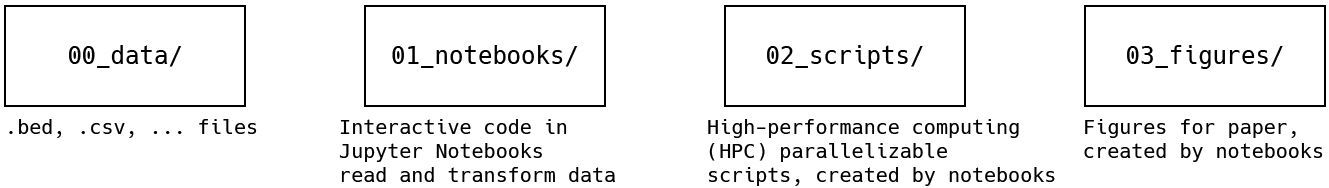This is the README.md file for code accompanying the paper, "Single-cell alternative splicing analysis with Expedition reveals splicing dynamics during neuron differentiation." by Song and Botvinnik, et al, Molecular Cell (2017). The focus of this study was to investigate alternative splicing at the single-cell level, in the system of motor neuron differentiation and applied to a model of neurodegenerative disease.
Some abbreviations:
- The celltypes are:
- Induced pluripotent stem cells (iPSCs)
- Neural progenitor cells (NPCs)
- Motor neurons (MNs)
- Motor neurons subjected to sodium arsenite, a cellular stress. Aka stressed motor neurons (sMNs)
hg19- The genome build of the human genome used in this project- Alternative splicing types/modes
- Skipped exons (SE)
- Isoform 1:
exon1-exon3 - Isoform 2:
exon1-exon2-exon3
- Isoform 1:
- Mutually exclusive exons (MXE)
- Isoform 1:
exon1-exon3-exon4 - Isoform 2:
exon1-exon2-exon4
- Isoform 1:
- Skipped exons (SE)
- Percent spliced-in (Psi/$\Psi$)
- A value between 0 and 1 (mathematically, [0, 1]) which represents what percentage of transcripts use this particular isoform, versus another isoform, e.g. isoform1 vs isoform2.
- For example, for skipped exon events, with
exon1-exon2-exon3as the three possible exons, this value represents what percentage of transcripts useexon1-exon2-exon3versus ones that use eitherexon1-exon2-exon3ANDexon1-exon3
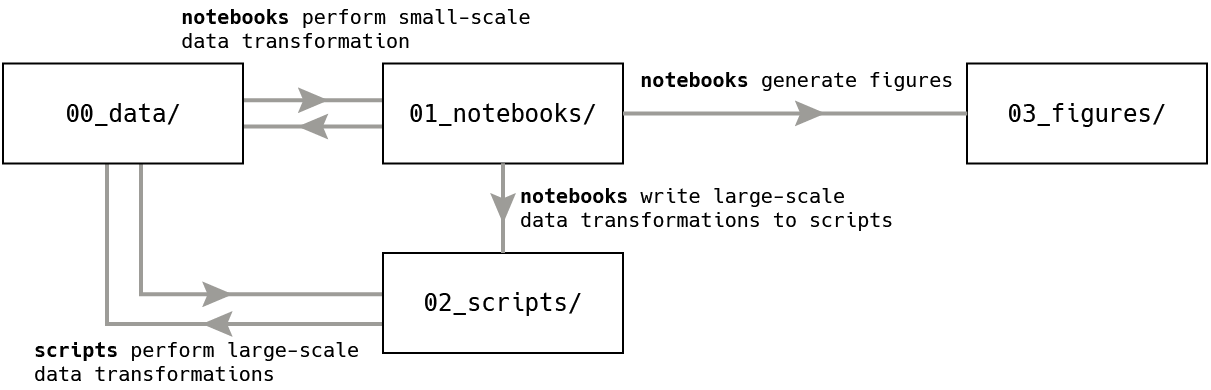
This repository has four main folders, numbered by the order in which the data flows through them:
00_data/- All input data files, after.bam(alignment), so.txt,.bed,.csvfiles01_notebooks/- Code to transformdatainto moredata, or writescriptsto transformdatainto moredatain a high-performance compute environment, or createfigures02_scripts/- Code written bynotebooksto use high performance computing to transformdatainto moredata03_figures/- Charts and plots for the paper
The organization of this repository is guided by the principles laid out by the
Reproducible Research Curriculum Hackathon -
Data Organization Subgroup,
where they recommend to organize a project's subfolders in the order that you
expect the information to flow, typically data → code → figures.
In this case, code represents both notebooks which were manually created to
explore the data do one of three things:
- Perform a transformation or filtering of the
data, if the transformation is not highly computationally intensive - Write a
scriptto submit to a high-performance compute clister to perform a transformation or filtering of thedata, if the transformation is highly computationally intensive - Generate a
figurefrom thedata
In case you're wondering, the font used in these diagrams is called Hack - thanks @freeman-lab!
Notebooks whose numbers begin with "0" are collecting data before any figures are created. Notebooks whose numbers begin 1-5 correspond to figures or data manipulation to do with that particular figure.
This repository is organized by the order of the figures in the paper. For
example the folder 01_Figure_1/ contains all the code (in IPython/Jupyter
Notebooks) to create the subpanels of Figure 1 and Supplementary Figure 1. The
exception is 00_Data_collection, which contains the most notebooks and
represents all the data collection, cleaning, and munging that needed to happen
so the figure making was easy.
Within each 0X_Figure_X/ folder, there is a subfolder called figures which
contains the graphs generated by the notebooks in the Figure folder.
Notebooks whose numbers begin with "0" are collecting data before any figures are created. Notebooks whose numbers begin 1-5 correspond to figures or data manipulation to do with that particular figure.
To ensure posterity of results, this document shows the versions of all software used in this paper: Python, Perl, and command line tools.
This notebook reads the sailfish and STAR output files to create gene expression and mapping stats matrices, and splice junction files. This also subsets the data separately on iPSC, NPC, and MN samples (Figures 1-4) and saves the stressed motor neurons (sMNs for Figure 5) separately.
A key aspect of this study is the annotation of alternative splicing events. We calcluate alternative splicing of skipped exon and mutually exclusive exons, based on read which uniquely mapped to a junction between exons.
As this is a large endeavor, the annotation steps are broken down into the following notebooks (single digits are prefixed by "0" for sorting purposes):
To calculate alternative splicing, outrigger does the following:
- Get annotated exons corresponding to splice junctions
- Identify all annotated (Gencode v19 - all levels) exons which are exactly upstream and downstream of all junctions.
- Create (junction, direction, exon) triples for storage in a graph database using
graphlite
- Create SE, MXE, and constitutive exon annotations de novo from (1)
- Aggregate exons and junctions into de novo splicing annotations of skipped exon (SE) and mutually exclusive exon (MXE) events.
- Calculate percent spliced-in (Psi) scores for MXE and SE events from (2)
- Calculate percent spliced-in (Psi/$\Psi$) of SE and MXE events, based on junction reads.
- Splicing gene id, name annotations with event definitions from (2)
- Start a splicing event annotation table.
- Get gene ids corresponding to SE and MXE event annotations
- Use these ids to intersect with previously created gene annotations.
After outrigger, the detected exons could have many properties and were found
using the following methods:
- Extract constitutive and alternative exons, write to bed file, submit job to
calculate conservation
- Create bed files of alternative and constitutive exons.
- Submit a compute job to calculate mean placental mammal conservation of
exon bodies (
bigWigAverageOverBed).
- Calculate exon properties (conservation, splice site strength) 2. Use bed files from (5) to calculate splice site strength (Yeo et al, J Comput Biol 2004 3. Read calcualted conservation from (5) 4. Add this information back to the splicing event annotation table.
- Ancient alternatively spliced exons (Merkin et al, 2012)
- Obtain ancient alternatively spliced exon table from Merkin et al, Science 2012
- Use
liftOverto convert rhesus macaque (rheMac2) coordinates to human (hg19) coordinates - Overlap with identified alternative exons from (2).
- Add this information back to the splicing event annotation table.
- Save conservation wiggle as a memory-mapped Genomic Array
- Use
HTSeqto save a 79 gigabyte memory-mapped file of the placental mammal conservation at every genomic base of the human genome. - This is done because it takes a long time to populate the array (overnight on a compute node) and it's easier to read in the pickle file, which takes about 30 min.
- Use
- Create CSVs of base-wise conservation of alternative and constitutive introns
- Read the pickled memory-mapped conservation file from (8)
- Get base-wise conservation of introns flanking alternative and constitutive exons. This will be used in Figure 2 when describing intron conservation of alternative splicing modalities.
- Transcribe isoforms to get RNA sequence
- Submit miRNA finding compute job on sequences from (10)
- Use first 17bp of microRNAs from mirbase, since the ends of them are bound by Ago
- Use
RNAhybridto estimate which microRNAs may bind an isoform's transcript
- Properties of the isoforms' RNA sequence from (10)
- Use BioPython to calculate...
- GC content
- maybe: [Estiamte RNA structure (single or double stranded)]
- Add this information back to the splicing event annotation table.
- Use BioPython to calculate...
- Translate isoforms to protein products
- Submit compute job for
hmmscanon Pfam-A, calculate disordered scores on translations from (13)- Use
hmmscanto find protein domains on the isoform translations from (13) - Use
IUPredto calculate average disordered region score of each isoform
- Use
- Calculate properties of isoform protein products (isoelectric point, etc) on translations from (13)
- Properties calculated:
- Molecular weight
- Isoelectric point
- ...
- Read disordered scores from (14)
- Add this information back to the splicing event annotation table.
- Properties calculated:
- Read
hmmscanoutput from (14) and get domain switching events- Aggregate domains on pfam clans to see whether the entire concept of a protein's function has changed
The gene annotations are less comprehensive and can be found in a single notebook. The steps are:
- Get gene id, name, level, biotype from
gtffile - Calculate maximum number of exons per gene
- Intersect with annotations from Gerstberger et al, Nat Rev Genet (2014)
- Transcription factors (TFs)
- Really defined as any RNA biologist would define TFs: "stuff that binds DNA" - i.e. proteins which induce transcription versus chromatin remodelers vs topoisomerases are all included in this group.
- RNA binding proteins, which target ...
- mRNA
- snRNA
- tRNA
- ribosome
- Transcription factors (TFs)
- Intersect with annotations from Animal TFDB
- Transcription factors
- Chromatin remodelers
- Transcription co-factors
- Intersect with Phylostratum annotations from Domazet-Loso and Tautz, Mol Biol Evol (2008)
- This gives the "age" of the gene, relative to the origin of life
- Intersect with genes associated with the cell cycle from Gene Ontology
A figure to make the reader feel "warm and fuzzy" - show that our celltypes express the marker genes that they are expected to, and alternative splicing specific to the celltypes.
Introduces a new approach to categorizing alternative splicing events (really, any percent-based units) by grouping them into "modalities."
- Get splicing events with at least 20 single cell observations per celltype
- Estimate per-cellytpe modality for each splicing event using
modish
- Investigate enrichment of properties from (0.2.*) in each modality
- Subset the bed files created in 0.2.5 to get each (phenotype, modality) subset
- Submit a compute job to find enrichment of intron motifs in (phenotype, modality) vs (phenotype, other modalities) using HOMER
- Change in modalities between celltypes
- Venn diagrams of modalities
- Heatmap/checkerboard of how modalities change from one celltype to the next
- Gene Ontology (GO) enrichment of modaliites
- (phenotype, modality) vs (phenotype, other modalities)
- (phenotype, modality) vs (other phenotype, modality)
- (modality in any phenotype) vs (other modality in any phenotype)
i.e. "What GO categories are iPSC bimodal events enriched for, compared to other modalities in iPSC?"
i.e. "What GO categories are iPSC bimodal events enriched for, compared to bimodal events in NPC and MN?"
i.e. "What GO categories are bimodal events in iPSC, NPC and MN enriched for, over other modalities in those celltypes?"
- Basewise conservation of flanking introns of modality exons.
- Use basewise conservation CSVs created in (0.2.09)
Changes in alternative splicing using change in modality and principal component analysis (PCA) on splicing events residing in constitutively expressed genes.
Introduces a new approach to visualizing and finding changing splicing events using bonvoyage to create a "Voyage Space"
Application of changing splicing events from modality finding using modish, voyaging events from bon voyage, and PCA on constitutively expressed, to the model of neurodegenerative disease of cellularly stressed motor neurons.