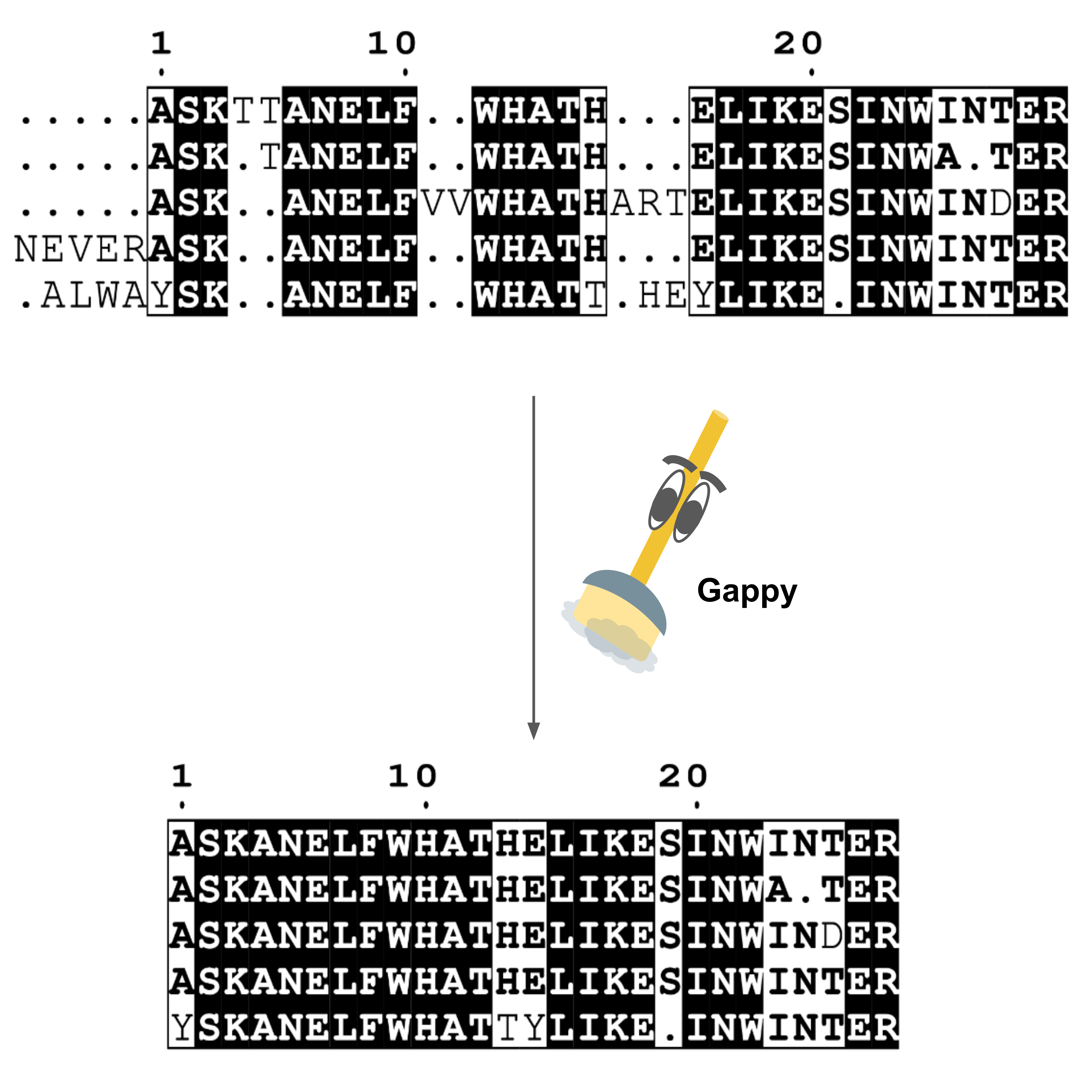
GapClean takes a gappy multiple sequence alignment and removes columns with gaps at a
specified threshold value to produce a "cleaner" and easier to visualize sequence alignment.
Can also be used to remove gaps in an alignment relative to a seed sequence to assess site-wise mutational information.
-i Input file (Required)
-o Output file (Required)
-t Threshold value (Optional) Cannot be used with seed argument
-s Seed index (Optional) Cannot be used with threshold argument
-h Display this help message
Example: gapclean -i input.fa -o output.fa -s 0 # Takes first sequence as the seed and removes gaps relative to seed.
-
Download gapclean and the bucket folder and copy both to your local bin (EG:
/usr/local/bin/) -
chmod +x gapclean -
Thank Gappy for his service. He is a retired detective.