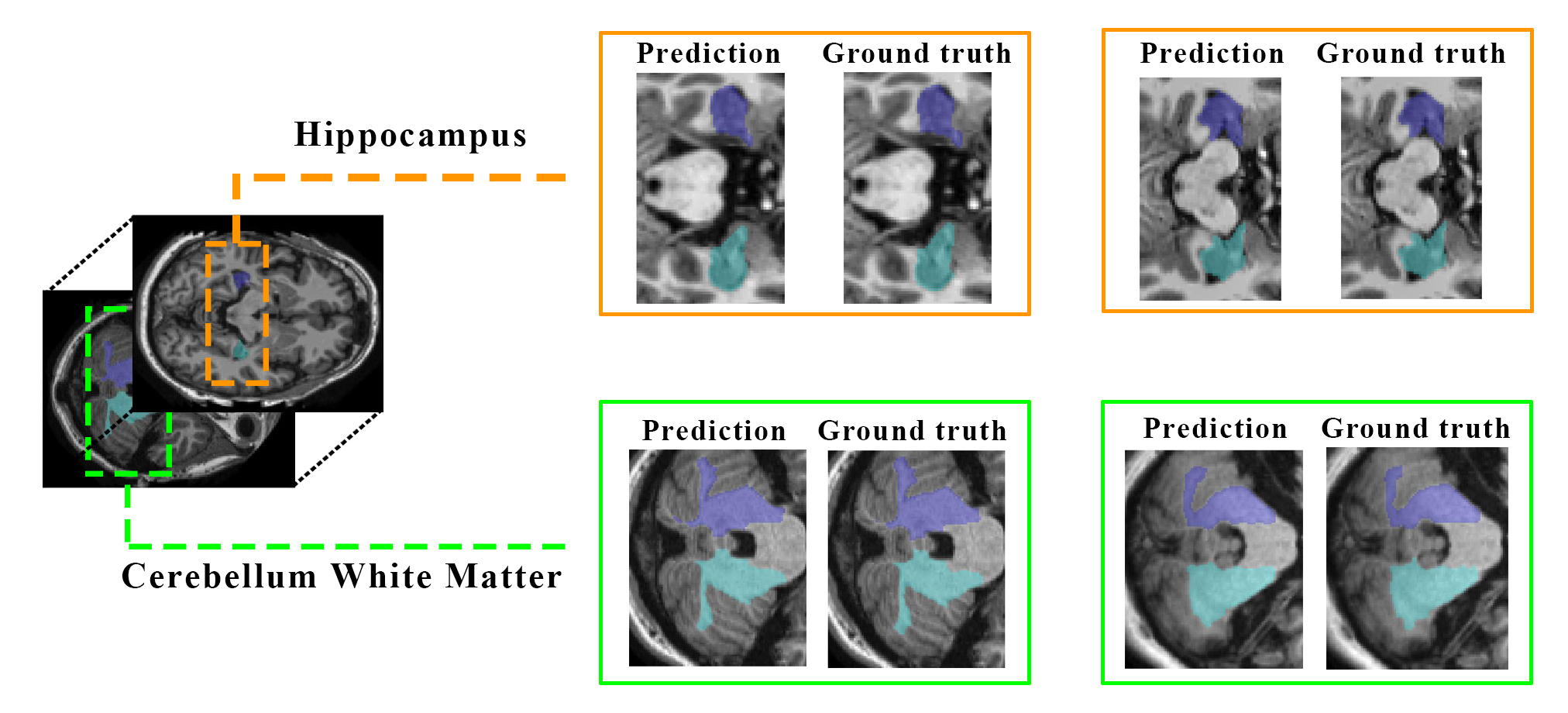
This code is an implementation of our proposed FLBS framework (IEEEXplore) in EMBC 2022 for brain segmentation for clinical use. For this reason our approach works with the minimum GPU requirements and reasonable time.
The code is written and tested on UBUNTU 18.06. For running the code you need:
- MATLAB
- nnU-Net dependency
For installing nnU-Net please follow the instruction on nnU-Net repository. Note that there is a step for configuration of the nnU-Net that should not be skiped.
The framework consists three main stages. For having the final results these parts should be executed in order. Before running the code, please customize the config.m file based on your own requirements.
In this part, the images dataset is registerd to MNI305 space and the intensities are normalized. For execution run preprocess.m file. The results will apear in temp file in code directory.
In case of already having the trained weights for nnU-Net, you can go straight to prediction. For the list of structures in config.m please contact us to provide you with the trained networks (final weights). For contact detail look at the end of this page.
For training using the prepared data run the training command defined on nnU-Net for each structure.
When pre-process stage in carried out, a command script will appaer in tmp folder. By executing the script in terminal or command line, the framework computes thepredictions.
bash predCommand.sh
Post-process merges all the predictions into a single segmentation map. Run the postprocess.m file on MATLAB. The restuls will appear in res folder in code directory. This is the final result.
In case of using this code in your research or project please cite the paper below:
A. Nejad, S. Masoudnia and M. -R. Nazem-Zadeh, "A Fast and Memory-Efficient Brain MRI Segmentation Framework for Clinical Applications," 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2022, pp. 2140-2143, doi: 10.1109/EMBC48229.2022.9871715.
This code has been implemented at Research Center for Science and Technology in Medicine, Tehran University of Medical Sciences, Tehran, Iran.
This project was funded by Iran's National Elites Foundation (بنیاد ملی نخبگان).