A journal club for -omicsy things @ UCLA. Send me a paper if you'd like to suggest one, otherwise I will choose.
We meet in the sixth floor conference room of Gonda Thursdays at 5:30 for beer and science. If you're reading this, feel free to stop by.
We are modeled after Jeff Ross-Ibarra's REHAB, please check there for information on the format. In short, no one presents the paper and we jump in as soon as the beer arrives.
We have a mailing list here. Feel free to join or email me (firstname.lastname@gmail.com) for paper annoucements.
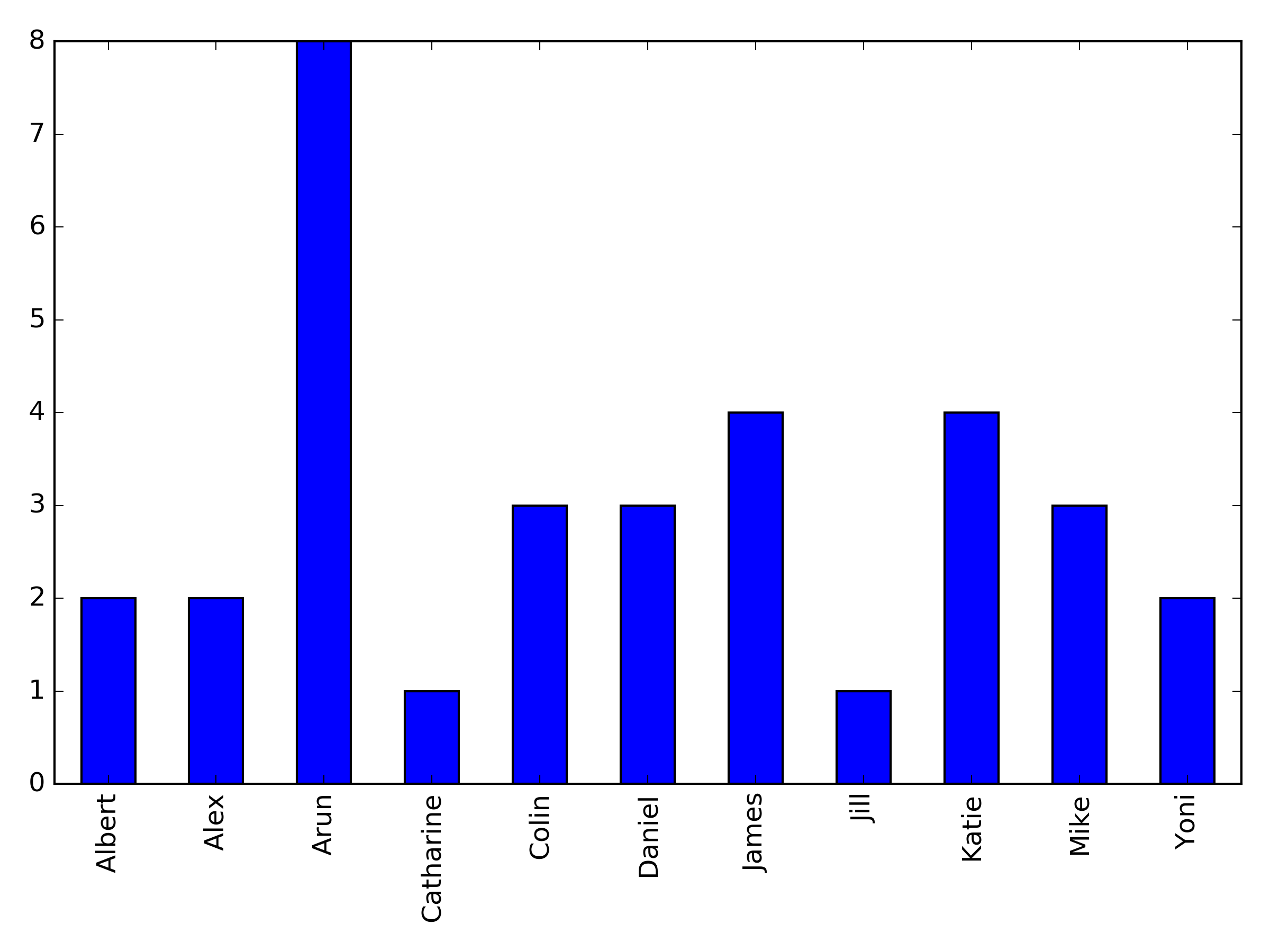
Here's some data on who's suggesting papers:
September 5th, 2019 Stuart et al 2019 Comprehensive integration of single-cell data [blame Eric]
May 16th, 2019 Border et al 2019 No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples [blame Katie]
April 18th, 2019 Wainschtein et al preprint 2019 Recovery of trait heritability from whole genome sequence data [blame James]
April 4th, 2019 Hinch et al 2019 Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm [blame Adriana]
March 21st, 2019 Li et al 2018 Decoding the genomics of abdominal aortic aneurysm [blame Maria]
February 28th, 2019 Castel et al 2018 Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk [blame Yoni]
January 17th, 2019 Larsson et al 2019 Genomic encoding of transcriptional burst kinetics [blame Katie]
May 11th, 2018 Matz et al 2018 Potential and limits for rapid genetic adaptation to warming in a Great Barrier Reef coral [blame Katie]
May 4th, 2018 Veeramah et al 2018 Population genomic analysis of elongated skulls reveals extensive female-biased immigration in Early Medieval Bavaria [blame Katie]
April 27th, 2018 Small et al 2018 Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition [blame Katie]
April 20th, 2018 Kremling et al 2018 Dysregulation of expression correlates with rare-allele burden and fitness loss in maize [blame Arun]
April 13th, 2018 Chen et al 2018 The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases [blame Katie]
March 16th, 2018 Alpert Sugden et al 2018 Localization of adaptive variants in human genomes using averaged one-dependence estimation [blame Daniel]
March 9th, 2018 Danko et al 2018 Dynamic evolution of regulatory element ensembles in primate CD4+ T cells [blame Arun]
March 2nd, 2018 No journal club -- go to UCLA Computational Genomics Winter Institute instead.
February 23rd, 2018 Lake et al 2018 Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain [blame Jill]
February 9th, 2018 Combs et al 2017 Spatial population genomics of the brown rat (Rattus norvegicus) in New York City [blame Katie]
February 2nd, 2018 Kong et al 2018 The nature of nurture: Effects of parental genotypes [blame Yoni]
January 26th, 2018 Hernandez et al 2017 Singleton Variants Dominate the Genetic Architecture of Human Gene Expression [blame Arun]
January 12th, 2018 Mumbach, Satpathy, Boyle and Dai et al 2017 Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements [blame Mike]
December 8th, 2017 Houshdaran et al 2016 Aberrant Endometrial DNA Methylome and Associated Gene Expression in Women with Endometriosis [blame Katie]
November 17th, 2017 Angermueller et al 2017 DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning [blame Colin]
November 3rd, 2017 Raj et al 2017 Integrative analyses of splicing in the aging brain: role in susceptibility to Alzheimer's Disease [blame Yoni]
October 27th, 2017 Ongen et al 2017. Estimating the causal tissues for complex traits and diseases [blame Arun]
October 20th, 2017 He et al 2017. Unified Sequence-Based Association Tests Allowing for Multiple Functional Annotations and Meta-analysis of Noncoding Variation in Metabochip Data [blame Albert]
October 13th, 2017 Weissensteiner et al 2017. Combination of short-read, long-read, and optical mapping assemblies reveals large-scale tandem repeat arrays with population genetic implications [blame Daniel]
October 6th, 2017 Rubin, Barajas, and Furlan-Magaril et al 2017. Lineage-specific dynamic and pre-established enhancer–promoter contacts cooperate in terminal differentiation [blame James]
September 29th, 2017 Lareau and Aryee 2017. diffloop: a computational framework for identifying and analyzing differential DNA loops from sequencing data [blame Katie]
September 21st, 2017 Castel et al 2017. Modified penetrance of coding variants by cis-regulatory variation shapes human traits [blame James]
September 15th, 2017 Mostafavi et al 2017. Identifying genetic variants that affect viability in large cohorts [blame Arun]
August 4th, 2017 Cortes and Dendrou et al 2017. Bayesian analysis of genetic association across tree-structured routine healthcare data in the UK Biobank [blame Colin]
No journal club -- go to UCLA Computational Genomics Summer Institute instead.
June 30th, 2017 Hammerschlag et al 2017. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits [blame Albert]
June 23rd, 2017 Boyle et al 2017. An Expanded View of Complex Traits: From Polygenic to Omnigenic [blame Catharine, James, Arun]
- Bonus: Goldstein 2009. Common Genetic Variation and Human Traits [blame James, Arun]
- Bonus: Rockman 2012. The QTN program and the alleles that matter for evolution: all that's gold does not glitter [blame Arun]
June 16th, 2017 McLaughlin et al 2017. Genetic correlation between amyotrophic lateral sclerosis and schizophrenia [blame Alex]
June 9th, 2017 Glodzik et al 2017. A somatic-mutational process recurrently duplicates germline susceptibility loci and tissue-specific super-enhancers in breast cancers [blame Mike]
June 2nd, 2017 Knouse et al 2016. Assessment of megabase-scale somatic copy number variation using single cell sequencing [blame Daniel]
May 25th, 2017 Mathieson et al 2015. Genome-wide patterns of selection in 230 ancient Eurasians [blame Arun] Note: date change
May 19th, 2017 No journal club -- go to Brewin' Talks!
May 12th, 2017 Pala and Zappala et al 2017. Population- and individual-specific regulatory variation in Sardinia [blame Arun]
May 5th, 2017 Corces et al 2017. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution [blame Mike]
April 28th, 2017 Saleheen & Natarajan et al 2017. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity [blame Alex]
April 21st, 2017 Yue et al 2017. Contrasting evolutionary genome dynamics between domesticated and wild yeasts [blame James]
April 14th, 2017: Chiang et al 2017. The impact of structural variation on human gene expression [blame Colin]
April 7th, 2017: Poplin et al 2016. Creating a universal SNP and small indel variant caller with deep neural networks [blame Arun]