This is a step-by-step guide showing how to use Chrom3D to generate a 3D genome model, and how to visalize this. The guide takes you through all steps from raw reads (fastq) to final results. To allow a relatively fast procedure, Hi-C input data have been downsampled to only include reads mapping to human chromosome 18. This tutural can be run on a personal computer (running MacOS or Linux) or an external server. The steps take ~30 minutes to run from start to end. A full-genome version of this tutorial is available here.
This tutorial uses HiC-Pro to map and process the Hi-C data into intra- and interchromosomal contact maps. From these, TADs are called using Armatus, which are used to delineate the genome into contiguous genome segments defining beads in the 3D genome models. Statistically significant interactions are identified between segments using the Non-central Hypergeometric distribution defining inter-bead constraints. LADs from chromatin immunoprecipitation-sequencing (ChIP-seq) of lamin B1 are used to restrain beads towards the nuclear periphery. The process of calling LADs from ChIP-seq reads is not considered here. The Hi-C and ChIP-seq data used here come from a published study of 4D genome dynamics during adipogenic differentiation. To generate the 3D genome model, we use Chrom3D. Chrom3D considers (i) significant pairwise interaction between genomic segments determined from Hi-C data to infer a 3D placement of these relative to each other, and (ii) significant interactions of chromatin with the nuclear lamina (LADs) determined e.g. from lamin ChIP-seq to infer a positioning of genomic segments relative to the nuclear periphery. In Chrom3D, chromosomes are modeled as chains of contiguous beads each representing a genome segment. The nucleus boundary is modeled as a sphere of 5 μm radius. Modeling starts from a random configuration of the chromosomes which is optimized using Monte Carlo simulations to produce a 3D genome structure. Reiterating the process hundreds of times, each time starting from a random bead configuration, yields an ensemble of 3D structures that recapitulate key features of 3D genome topologies in a population of cells. The final 3D models are visualized using UCSF ChimeraX.
To perform the steps in this step-by-step guide a computer running Unix/Linux or macOS, and/or a server running Linux is needed. The bash shell is used to run all the commands specified in the step-by-step guide. It is assumed that the user is somewhat knowledgeable in using the command line.
To follow the steps in this tutorial you will first need to install some software (see list below). Some of these tools might already be installed on your system, and other tools might be a bit challenging to install, such as Armatus and HiC-Pro. If you are having trouble installing the tools, ask your system administrator or a colleague with knownedge of UNIX systems to help you.
- Boost (Installation instructions) (required by Chrom3D and Armatus)
- Chrom3D v1.0.2 (Installation instructions) (available in your path)
- Armatus 2.3 (Installation instructions) (available in your path)
- HiC-Pro 2.11 (Installation instructions) (available in your path)
- Bedtools2 (Installation instructions) (available in your path)
- Python 2.7 or 3 (Installation instructions) (available in your path)
- statsmodels (Installation instructions)
- ChimeraX (Installation instructions) (installed on your local computer)
A step-by-step guide showing how Chrom3D can be used to generate a 3D genome model of human chromsome 18
1. Setup this tutorial and download the Hi-C data
git clone https://github.com/Chrom3D/INC-tutorial.git
cd INC-tutorial2. Download LAD data*
We will use LADs (in BED format) based on Lamin B1 ChIP-seq data in the same cell-line.
mkdir lad
curl -o lad/GSE109924_lad_D0-rep1.bed.gz https://ftp.ncbi.nlm.nih.gov/geo/series/GSE109nnn/GSE109924/suppl/GSE109924_lad_D0-rep1.bed.gz
gunzip lad/GSE109924_lad_D0-rep1.bed.gz3. Setting up HiC-Pro
HiC-Pro will be used to process the Hi-C data, including mapping the reads and aggregation of the contact frequencies (takes a few minutes).
curl -O https://raw.githubusercontent.com/nservant/HiC-Pro/master/config-hicpro.txt
curl -O https://raw.githubusercontent.com/nservant/HiC-Pro/master/annotation/chrom_hg19.sizes
curl -O https://raw.githubusercontent.com/nservant/HiC-Pro/master/annotation/HindIII_resfrag_hg19.bed
tar -zxvf hg19_chr18/* -C hg19_chr184. Downloading and installing the required processing scripts*
As part of the Chrom3D package, several processing scripts are available to reformat and process the data to prepare for running Chrom3D.
mkdir processing_scripts
curl -OL https://github.com/Chrom3D/preprocess_scripts/archive/v.1.2.zip
unzip -j -d processing_scripts/ v.1.2.zip preprocess_scripts-v.1.2/NCHG_hic.zip
unzip -j -d processing_scripts/ v.1.2.zip preprocess_scripts-v.1.2/cap_chr_end.py
unzip -j -d processing_scripts/ v.1.2.zip preprocess_scripts-v.1.2/make_diploid_gtrack.py
unzip -j -d processing_scripts/ v.1.2.zip preprocess_scripts-v.1.2/NCHG_fdr_oddratio_calc.py
unzip -j -d processing_scripts/ v.1.2.zip preprocess_scripts-v.1.2/unmappable_blacklist.bed
unzip -j -d processing_scripts/ v.1.2.zip preprocess_scripts-v.1.2/makeGtrack.py5. Installing NCHG
This program, which is also part of the Chrom3D package, determines statistically significant interactions from Hi-C data based on the Non-central Hypergeometric distribution.
unzip -d processing_scripts/ processing_scripts/NCHG_hic.zip
make -C processing_scripts/NCHG_hic/6. Adapting the config-hicpro.txt file
To prepare for running HiC-Pro, we will need to change two lines in the config-hicpro.txt file. Use a text-editor (like emacs, vim, nano, (or TextEdit [MacOS]) to:
- Add the bowtie path to line nr. 39:
BOWTIE2_IDX_PATH =->BOWTIE2_IDX_PATH = [fullpath]/hg19_chr18/where[fullpath]is the full path to your current working directory, i.e. theINC-tutorialdirectory (use thepwdcommand if you are uncertain about the working dir.) - Change the reference genome on 47:
REFERENCE_GENOME =->REFERENCE_GENOME = hg19_chr18 - Change line nr. 89:
BIN_SIZE = 20000 40000 150000 500000 1000000->BIN_SIZE = 50000 1000000
7. Run HiC-Pro (Takes ~10 minutes)
HiC-Pro --input fastq --output hicpro_results --conf config-hicpro.txt! If you are stuck at this point, you can copy nesessary files to proceed with the remaining steps by:
mkdir -p hicpro_results/hic_results/matrix/chr18/raw/50000/
cp backup/7/* hicpro_results/hic_results/matrix/chr18/raw/50000/8. Setup the folder structure for the HiC contacts
mkdir -p hic/bedpe/intra
mkdir -p hic/matrix9. Convert Hi-C to BEDPE and matrix format*
The output from HiC-Pro needs to be converte to BEDPE in order to be processed further by Chrom3D, and to a matrix format in order to be compatible with the Armatus TAD caller.
awk 'NR==FNR { map[$4] = $1"\t"$2"\t"$3; next } { print $0,map[$1],map[$2] }' hicpro_results/hic_results/matrix/chr18/raw/50000/chr18_50000_abs.bed hicpro_results/hic_results/matrix/chr18/raw/50000/chr18_50000.matrix | awk '$4==$7' | awk '{print $4"\t"$5"\t"$6"\t"$7"\t"$8"\t"$9"\t"$3}' > hic/bedpe/intra/chr18
for chr in hic/bedpe/intra/*
do
chrname=$(basename $chr)
cut -f 2,5,7 $chr > hic/matrix/$chrname
done!If you are stuck at this point, you can copy nesessary files to proceed with the remaining steps by:
mkdir -p hic/bedpe/intra/
mkdir -p hic/matrix/
cp backup/9/bedpe/chr18 hic/bedpe/intra/
cp backup/9/matrix/chr18 hic/matrix/10. Running Armatus to call TADs
mkdir hic/tads
armatus-linux-x64 -r 50000 -c chr18 -S -i hic/matrix/chr18 -g .6 -o hic/tads/chr18Note: if you get error messages here, try to see if you have installed armatus such that you can execute it as armatus instead of armatus-linux-x64.
The -r 50000 sets the bin-size to 50000 bp, -c chr18 specifies that only chromosome 18 should be considdered, -S specifies that sparse matrix format (3 column text file) is used, -i hic/matrix/chr18 provides the input data (in matrix format), -g .6 is the gamma-max parameter indicating the highest resolution to generate domains (often is set based on trial and error), -o hic/tads/chr18 gives the output for the TADs.
!If you are stuck at this point, you can copy nesessary files to proceed with the remaining steps by:
mkdir -p hic/tads/
cp backup/10/chr18.consensus.txt hic/tads/11. Since Armatus output is end-inclusive, convert to BED by adding 1 to end position
awk '{printf("%s\t%i\t%i\n",$1,$2,$3+1)}' hic/tads/chr18.consensus.txt > hic/tads/chr18.consensus.bed12. Convert called TADs into a segmented genome to define Chrom3D beads
bedtools complement -L -i hic/tads/chr18.consensus.bed -g chrom_hg19.sizes | cat - hic/tads/chr18.consensus.bed | bedtools sort -g chrom_hg19.sizes > chr18_beads.bedThis one-liner combines the complementary (non-TAD) genomic regions (found using bedtools complement) with the TADs to generate a fully segmented chromosome 18 (needed by Chrom3D). -L in bedtools complement limits output to solely the chromosomes with records in the input file. -g specifies the chromosome size. bedtools sort orders the genomic regions by their position on the chromosome. This file will define the "beads" in the Chrom3D model.
13. Map intra-chromosomal interactions from Hi-C to the beads defined in the previous step and aggregate the contacts between these beads
cat hic/bedpe/intra/chr* | awk '{printf("%s\t%s\t%s\n",$1,$2,$2+1)}' | bedtools intersect -wao -a stdin -b chr18_beads.bed | cut -f 4,5,6 > left.tmp
cat hic/bedpe/intra/chr* | awk '{printf("%s\t%s\t%s\t%s\n",$4,$5,$5+1,$7)}' | bedtools intersect -wao -a stdin -b chr18_beads.bed | awk '{printf("%s\t%s\t%s\t%s\n",$5,$6,$7,$4)}' > right.tmp
paste left.tmp right.tmp | awk '{a[$1"\t"$2"\t"$3"\t"$4"\t"$5"\t"$6] += $7} END{for (i in a) print i"\t"a[i]}' | awk '$2!=$5' | sort -k 2n,2n > chr18_bead_interactions.intra.bedpe
rm left.tmp right.tmpThis maps the left and right part of the Hi-C interactions to the using bedtools intersect to the Chrom3D beads defined in the previous step. Then, the left and right parts are combined (with paste) and their Hi-C contact frequencies are aggregated (using awk) to generate a BEDPE file consisting of contact frequencies between all beads on the chromosome.
14. Remove interactions between beads overlapping centromeres
curl -s "http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/cytoBand.txt.gz" | gunzip -c | grep acen | bedtools pairtobed -a chr18_bead_interactions.intra.bedpe -b stdin -type neither > chr18_bead_interactions.intra.nocen.bedpeDownload (using curl) a textfile containing information about cytobands in hg19, then unzip this file and fetch all centromeric regions ("acen"). These regions are then overlapped with the chr18 beads positions using bedtools pairtobed. With bedtools pairtobed using -type neither, only interactions where neither of the beads overlap with centromeres are retained. This is done, since interactions involving centromeric beads are often artefactual.
!If you are stuck at this point, you can copy nesessary files to proceed with the remaining steps by:
cp backup/14/chr18_bead_interactions.intra.nocen.bedpe .15. Identifying statistically significant inter-bead interactions within chromosomes, using the Non-central Hypergeometric distribution (NCHG)
processing_scripts/NCHG_hic/NCHG -m 50000 -p chr18_bead_interactions.intra.nocen.bedpe > chr18_bead_interactions.intra.nocen.NCHG.out
python processing_scripts/NCHG_fdr_oddratio_calc.py chr18_bead_interactions.intra.nocen.NCHG.out fdr_bh 2 0.01 > chr18_bead_interactions.intra.nocen.NCHG.sigNote: if python processing_scripts/NCHG_fdr_oddratio_calc.py gives you error messages, try to replace python with python3 or python2.7 (depending on where statsmodels was installed)
Here, NCHG is used to determine statistically singificant interactions between beads. -m 50000 gives the minimum interaction length (in bp), -p chr18_bead_interactions.intra.nocen.bedpe specifies the input file. The output is redirected into the file chr18_bead_interactions.intra.nocen.NCHG.sig. The NCHG program uses the Non-central Hypergeometric distribution to determine statistically singificant interactions. This model takes into account the total number of interactions on the chromosome, the number of interactions for the two involved beads, and the genomic/linear distance between the two beads. The processing_scripts/NCHG_fdr_oddratio_calc.py script is used to select significant interactions after multiple testing correction using the Benjamini-Hochberg method (fdr_bh) at the 0.01 level, and with an overserved/expected ratio of 2.
!If you are stuck at this point, you can copy nesessary files to proceed with the remaining steps by:
cp backup/15/chr18_bead_interactions.intra.nocen.NCHG.sig .16. Generate the Chrom3D input file in GTrack format, specifying the 3D model setup
Chrom3D relies on the GTrack file format for specifying the model setup. This includes the genomic position of the beads, their size, interactions between them and interactions with the nuclear periphery. This input file also can be used to define the color of the beads.
python processing_scripts/makeGtrack.py chr18_bead_interactions.intra.nocen.NCHG.sig chr18_beads.bed > chr18_bead_interactions.gtrack
echo -e "##gtrack version: 1.0\n##track type: linked segments\n###seqid\tstart\tend\tid\tradius\tperiphery\tedges" > chr18_bead_interactions.lads.gtrack
bedtools intersect -c -a chr18_bead_interactions.gtrack -b lad/GSE109924_lad_D0-rep1.bed | awk '{if($7>=1) print $1 "\t" $2 "\t" $3 "\t" $4 "\t" $5 "\t1\t" $6; else print $1 "\t" $2 "\t" $3 "\t" $4 "\t" $5 "\t.\t" $6}' >> chr18_bead_interactions.lads.gtrack The script processing_scripts/makeGtrack.py is used to define all beads in chr18_beads.bed in GTrack format, and specifies interactions between these beads based on the chr18_bead_interactions.intra.nocen.NCHG.sig file. Then, the GTrack header is added (echo -e...) before bedtools intersect -c is used to overlap each bead defined in the GTrack file with lads defined in the lad/GSE109924_lad_D0-rep1.bed file. Note that the GTrack file is treated as a regular BED file by bedtools, illustrating the convenience of the GTrack format in combinaton with bedtools.
!If you are stuck at this point, you can copy nesessary files to proceed with the remaining steps by:
cp backup/16/chr18_bead_interactions.lads.gtrack .17. Run Chrom3D based on the GTrack file
Chrom3D -n 100000 -r 5.0 --nucleus -y 0.01 -l 10000 -c 0.001 chr18_bead_interactions.lads.gtrack > model.cmmThis step runs Chrom3D according to the model options defined in the chr18_bead_interactions.lads.gtrack file.
The parameter -n 100000 specifies the total number of iterations (100,000), -r 5.0 sets the nuclear radius to 5μm, --nucleus specififies that all beads should be constrained within the nucleus, -y 0.01 sets the volume of the chromosome model to be 1% of the total volume of the nucleus, -l 10000 specifies that logging information should be output every 10,000 iterations. The final parameter -c 0.001 sets the cooling-rate. When this number is set different from 0 (like here) the optimization procedure uses simulated annealing. The temperature will decrease gradually during the simulation with the slope determined by c for each accepted move. The simulated annealing approach works better/faster in single chromosome models, whereas a Metropolis-Hastings (-c 0 [default]) criterion is better for larger systems consisting of multiple chromosomes.
!If you are stuck at this point, you can copy nesessary files to proceed with the remaining steps by:
cp backup/17/model.cmm
cp backup/17/model_redlad.cmm18. Visualizing model.cmm in ChimeraX
- If you are running this step-by-step guide on a server, download the
model.cmmfile to your local computer - The resulting
model.cmm(andmodel_redlad.cmmfrom step 19) can be opened in ChimeraX and displays of these turned on and off in the bottom right "Models" panel. To generate tomographic views of models, the commandclipcan be used in the "Command:" field in the bottom panel of ChimeraX. Background color and other graphical adjustments can be performed by clicking the "Graphics" button in the top panel. - In ChimeraX, the command
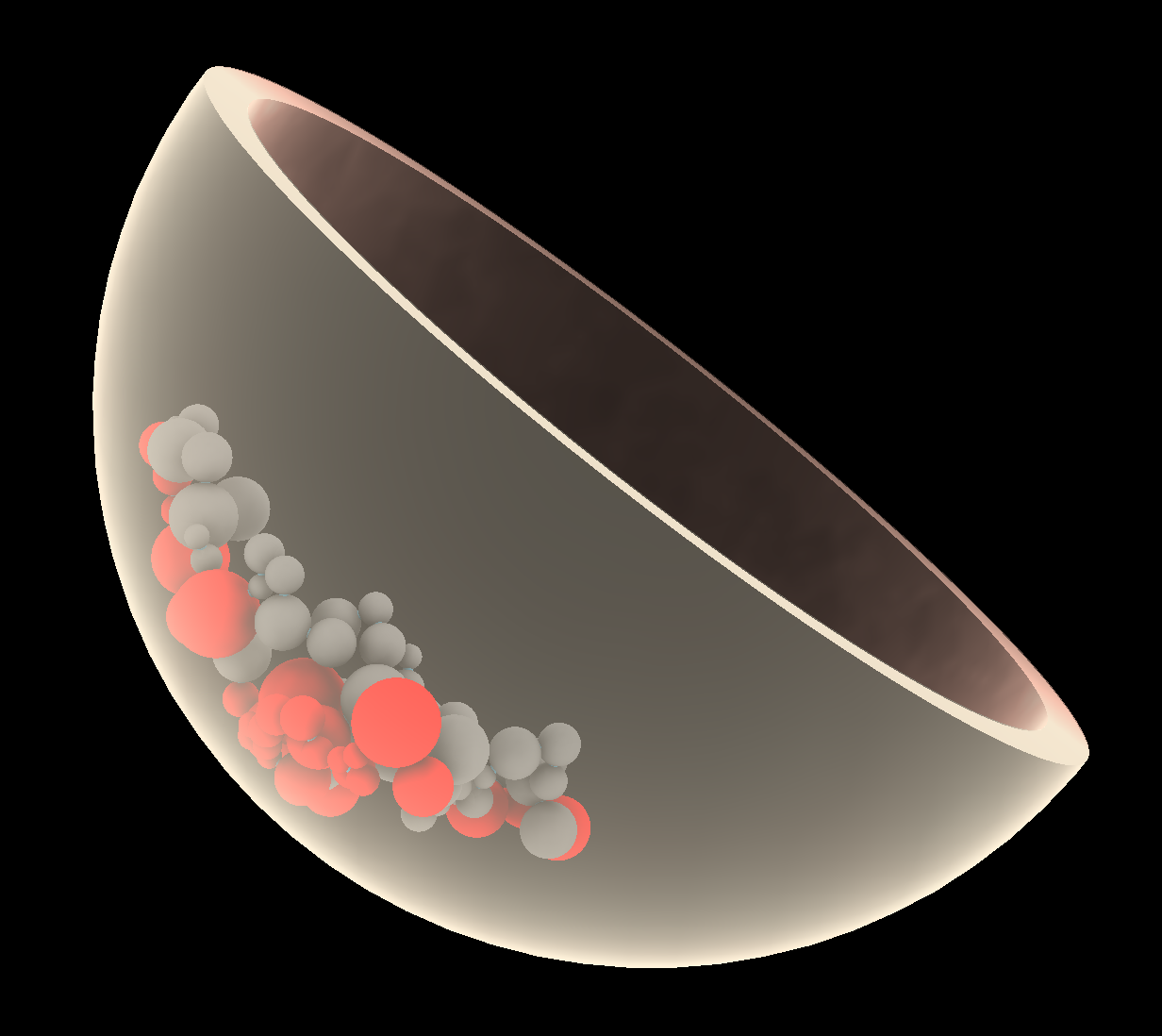
shape sphere center 0,0,0 radius 5.0 color #ffc9b5 slab 0.5can be used in the "Command" field in the bottom panel to display a nucleus structure on top of the model view. To change opacity of the nucleus model, click the colored square called "sphere" in the "Models" panel in the bottom right of the view, and select e.g. 30%. - Again, "clip" can be used to clip this to generate tomographic views. The model can also be tilted to allow a better perception of depth in the structures. Figure 4 shows some of the resulting illustrations that can be generated using ChimeraX.
- Tip: if you want white background, run the command
lighting depthCueColor blackbefore you select White in the Background section of the top panel - Tip: by running the command
shape sphere center 0,0,0 radius 5.0 color #ffc9b5 mesh true, you get a mesh/grid visualization of the nuclear boundary
19. Coloring beads defined by LADs using red color
unzip -j -d processing_scripts/ v.1.2.zip preprocess_scripts-v.1.2/color_beads.py
awk '$6==1' chr18_bead_interactions.lads.gtrack | cut -f 4 > lads.ids
python processing_scripts/color_beads.py model.cmm lads.ids 255,0,0 OVERRIDE > model_redlad.cmmThe lads.ids now contains the ids (4th column) of all the beads with a periphery constraint (6th column) of 1. The processing_scripts/color_beads.py script specifies that all beads with ids listed in lads.ids should be colored in RGB value 255,0,0 (i.e. red color) in the Chrom3D output file (CMM file format). The OVERRIDE keyword specifies that any existing color definitions in the file should be ignored.
- Do the steps from step 18., but open and visualize the
model_redlad.cmmfile instead
Example of visualization in ChimeraX:
*If you are completely stuck, redo the steps marked with an asterisk (*), and jump in on one of the orange (!) lines