Fast and convenient lowest common ancestor (LCA) assignment from command line or Python using One Codex and EBI APIs. Capable of ~100 requests/s with the wind blowing in the right direction.
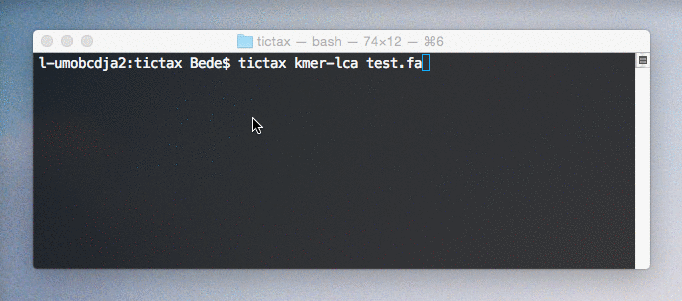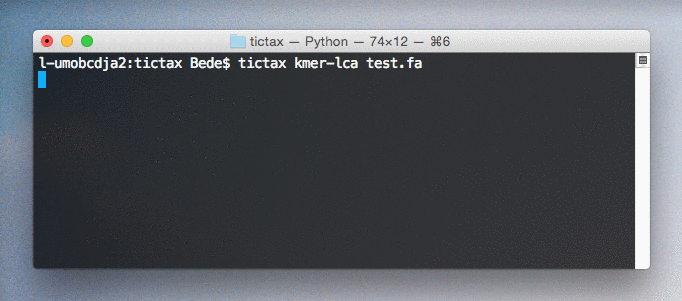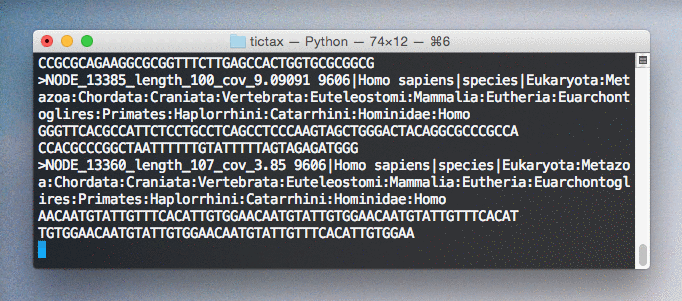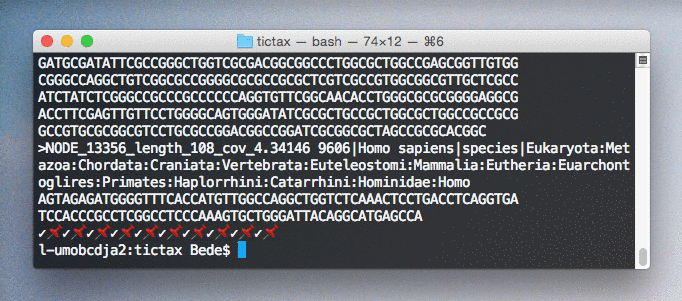
Scientific names and lineages are retrieved using the EBI taxonomy API and annotated within fasta description fields for easy viewing and parsing.
Tictax currently supports One Codex classification only. BLAST-based LCA (slow, sensitive) coming soon
$ tictax kmer-lca test.fa
$ tictax kmer-lca --progress test.fa > test.tt.fa
$ tictax -h
usage: tictax [-h] {kmer-lca} ...
positional arguments:
{kmer-lca}
kmer-lca Lowest common ancestor sequence assignment using the One Codex
API. Streams annotated records to stdout in fasta format. Taxa
assigned using the One Codex 31mer LCA database.
optional arguments:
-h, --help show this help message and exit
$ tictax kmer-lca -h
usage: tictax kmer-lca [-h] [-f] [-p] seqs-path
Lowest common ancestor sequence assignment using the One Codex API.
Streams annotated records to stdout in fasta format.
Taxa assigned using the One Codex 31mer LCA database.
positional arguments:
fasta-path path to fasta formatted input
optional arguments:
-h, --help show this help message and exit
-f, --fastq input is fastq; disable autodetection (default: False)
-p, --progress show progress bar (sent to stderr) (default: False)
- Returns Biopython SeqRecords with tictax annotations as the
descriptionattribute - LCAs are assigned using an LCA index of 31mers from the One Codex database
import tictax
records = tictax.parse_seqs('test.fa') # Biopython SeqRecord generator
records_classified = tictax.kmer_lca_records(records, one_codex_api_key) # List of SeqRecords
print(records_classified.format('fasta')) # Generate multifastaRequires Python 3.5+. Installs with pip.
pip3 install tictax
Feel free to open issues and PRs, else tweet or mail me via b àt bede dawt im.
-
blast-lcacommand - add BLAST LCA functionality - Switch to semicolon delimiters
- Add in onecodexrt / ebiblastn
-
kmer-lcacommand--outoption for two column id to LCA info -
abundancecommand for generating taxonomic abundances matrices at a specified rank - Stream input from stdin
- Use
aiohttp.errorsfor exception handling - Option to return records as async generator
- Support
fastqandgzipinput