NGS-MSTB: Next Generation Sequencing Microbial Surveillance Toolbox for scalable de-novo assembly of viral genomes and bacterial genes
This repository MICGENT is the main component of the NGS-MSTB system. Here we describe how to use the entire system that is comprised of multiple code repositories. If you want to skip the introduction, you can jump directly to running the NGS-MSTB as a Docker container.
NGS-MSTB is a bioinformatics pipeline for quickly and reliably assembling next generation sequencing reads from large scale viral and bacterial clinical strain surveillance studies to monitor potential drug resistance and the presence of virulence factors. Using the pipeline’s integrated graphical interface, an analyst can routinely assemble and review thousands of datasets in one batch within a few hours on a high-performance compute cluster.
The software assembles de-novo complete viral genomes from amplicon-derived shotgun reads potentially sequenced with extremely high local coverage variability as well as specific bacterial genes from whole genome shotgun reads.
The software was successfully deployed as the assembly module in a system validated according to good clinical practices (GCP). To this end, it guarantees deterministic results, full provenance data and ensures the integrity of the inputs and intermediate datasets through cryptographic checksums.
Source code, binary Conda packages and testing datasets are freely available from several associated repositories in GitHub NGS-MSTB organization and in Zenodo NGS-MSTB community. The pipeline provides a user interface based on the Galaxy bioinformatics workbench, dynamic Web reports with IGV.js genome browser views and a distributed backend coded with Common Workflow Language (CWL) and Python. CWL is executed by a locally modified Toil distributed workflow engine. A Docker container is available to simplify deployment. The software integrates the locally modified versions of ARIBA and Pilon. BBTools package is used in multiple places inside the pipeline. Spades is used as a key component in the assembly of viral datasets. Minimap2 is used for aligning assembly contigs to the reference.
You can view the high-level flow chart.
Andrey Tovchigrechko <andreyto AT gmail.com>
GPLv3. See also COPYING file that accompanies the source code.
The manuscript is currently under review:
A. Tovchigrechko, E. Aspinal, T. Slidel, H. Liu, B. Lu, A. Ruzin, R.J. Lebbink, H. Jin, M.E. Abram, M.T. Esser and D.E. Tabor, NGS-MSTB: Next Generation Sequencing Microbial Surveillance Toolbox for scalable de-novo assembly of viral genomes and bacterial genes.
In the meantime, you can cite the software release DOI on Zenodo:
Liu, Hui, Bin Lu, David E. Tabor, Andrey Tovchigrechko, Deidre Wilkins, Hong Jin, Shabir A. Madhi, Nasiha Soofie, Mark T. Esser, and Marta C. Nunes. "Characterization of human respiratory syncytial virus (RSV) isolated from HIV‐exposed‐uninfected and HIV‐unexposed infants in South Africa during 2015‐2017." Influenza and Other Respiratory Viruses (2020) https://doi.org/10.1111/irv.12727
David E. Tabor, Fiona Fernandes, Annefleur C. Langedijk, Deidre Wilkins, Robert Jan Lebbink, Andrey Tovchigrechko, Alexey Ruzin, Leyla Kragten-Tabatabaie, Hong Jin, Mark T. Esser, Louis J. Bont, Michael E. Abram, the INFORM-RSV Study Group, Global molecular epidemiology of RSV from the 2017-2018 INFORM-RSV study, Journal of Clinical Microbiology Oct 2020, JCM.01828-20; https://doi.org/10.1128/jcm.01828-20
Kurasawa, James H., Andrew Park, Carrie R. Sowers, Rebecca A. Halpin, Andrey Tovchigrechko, Claire L. Dobson, Albert E. Schmelzer, Changshou Gao, Susan D. Wilson, and Yasuhiro Ikeda. "Chemically-Defined, High-Density Insect Cell-Based Expression System for Scalable AAV Vector Production." Molecular Therapy-Methods & Clinical Development (2020) https://doi.org/10.1016/j.omtm.2020.09.018
David E. Tabor, Christine Tkaczyk, Andrey Tovchigrechko, Bret R. Sellman, Michael McCarthy, Pin Ren, Kathryn Shoemaker, Hasan S. Jafri, Bruno François, Mark T. Esser, Jasmine Coppens, Leen Timbermont, Basil Xavier, Christine Lammens, Herman Goossens, Surbhi Malhotra-Kumar, Alexey Ruzin, 1486. Phylogenetic and alpha toxin variant analyses of Staphylococcus aureus strains isolated from patients during the SAATELLITE study, Open Forum Infectious Diseases, Volume 7, Issue Supplement_1, Oct 2020, Pages S744–S745, https://doi.org/10.1093/ofid/ofaa439.1667
Unless you need a very fast turnaround (a few hours) on batches of thousands of sequencing datasets (samples), you will probably be OK with running our provided NGS-MSTB Docker image on a single multicore host machine. The machine can be a virtual machine (VM) in a commercial cloud, on-premise server or a laptop. Increasing the number of cores and, proportionally, RAM on the host machine will give you a roughly linear increase in performance over the number of samples. This is generally known as "vertical scaling". Using a Docker image greatly simplifies deployment of this complex integrated pipeline. For scaling beyond a single multicore machine, you will have to deploy the NGS-MSTB Conda packages and Galaxy tools into the existing Galaxy instance that is configured to run jobs on a distributed compute cluster comprising multiple nodes (horizontal scaling).
Currently, the dynamic Web report that NGS-MSTB generates will load several open-source JavaScript libraries from the public Content Distribution Networks (CDNs). This means that the client machine where you view the report in a Web browser will need access to the Internet.
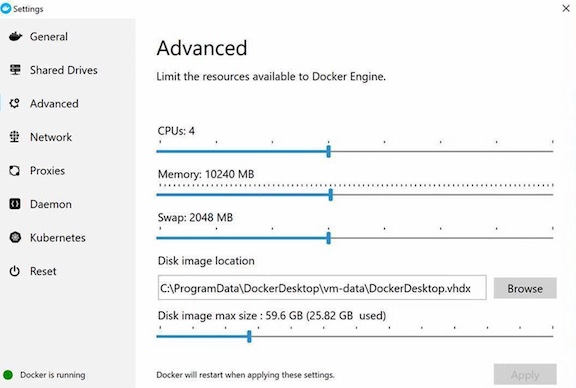
This assumes that you already have made some system operational on the host machine allowing you to run Docker containers. Our examples use Docker commands entered from a Linux terminal. You can easily adapt the commands to a different container execution system such as Redhat Podman and/or different host operating system (MacOS). Most of our testing of the NGS-MSTB container has been done on a Linux VM with 64GB RAM and 16 CPU cores and Mac PowerBook Pro with 32GB RAM and 6 CPU cores (12 cores with hyperthreading). On the Mac, we tested under minimum resources of 10GB RAM and 4 CPU cores made available to the Docker Desktop.
The Docker containers should be able to use at least four CPU cores and 10 GB of RAM. On Linux hosts, the containers by default have access to all CPUs and RAM of the host. On Mac, the containers run inside a VM, and the default Docker settings restrict the VM resource allocation. You should check and adjust these if necessary - the default settings may be too tight for NGS-MSTB. For example, under Docker Desktop, these settings will be found under Preferences -> Advanced. You should make and apply any changes to Docker Desktop before running the Docker commands because the Docker VM will get restarted and all running commands or containers terminated.
So far, we were not able to execute the full assembly workflow in Docker on Windows. On Windows, the workflow inevitably stalls with zero CPU usage and no tracable error messages despite all our attempts at adjusting the Docker resource parameters. This must be related to some subtle problems in the implementation of the Docker system itself on Windows OS. This might change in future versions of Docker or in Docker on Windows WSL 2. We will update this section if that happens. For now, NGS-MSTB should be used only on Linux or Mac hosts.
See below in this text details about the disk space requirements.
If you are increasing the hardware resources in order to process your data faster, make sure that you increment cores and RAM proportionally: for each four additional CPU cores, add eight GB of RAM.
If you are using any non-Linux host OS, make sure that you configure the VM that runs Docker
to use at least 10 GB of RAM, 4 CPU cores and have 10 GB of free disk space in the VM.
All of those must be configured in the Docker Desktop preferences or in the Virtual Box VM settings of
the docker-toolbox. The workflow will stall and never finish if it runs against the VM
disk space or swap limits.
docker run --rm -d -p 8080:80 --name ngs-mstb -e GALAXY_CONFIG_SINGLE_USER=admin_ge@ngs-mstb.nowhere ngsmstb/ngs-mstb:latest
In your Web browser, open the URL localhost:8080. If you started the container on a remote machine, replace localhost in the URL with the DNS name of that machine. You can also use a different port instead of 8080 both in the docker run command and in the URL. The recommended Web browser is Chrome, Firefox or other standards-complying browser.
The Galaxy instance inside the container will take about a minute to become fully operational - you might see Connection refused when you first type the URL and have to refresh the page a couple of times. Eventually, you will see the Galaxy user interface (UI) in your browser window.
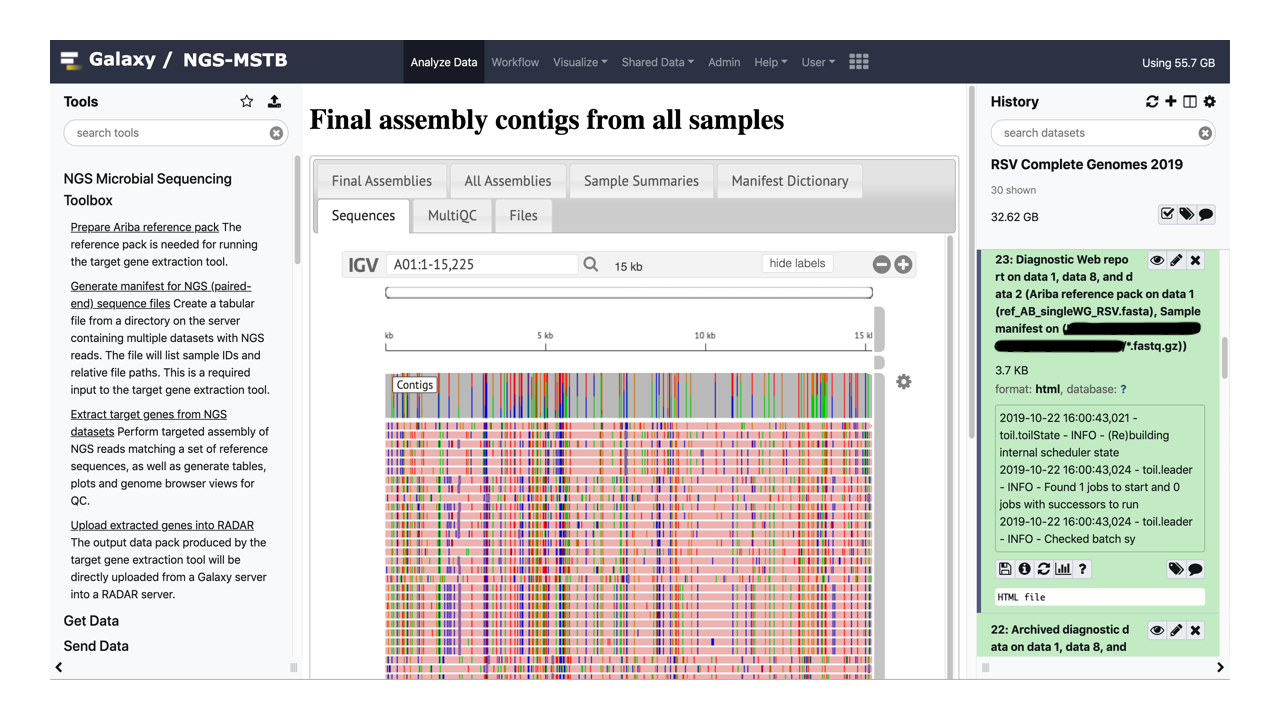
Follow the links at the center of the Galaxy front page to take the interactive tours of executing the example of assembling several complete genomes of Respiratory Syncytial Virus (RSV) and reviewing the resulting Web report. The tours allow comparing the NGS-MSTB assemblies of several NGS replicates against the independently obtained Sanger-based assemblies of the same genomes. All necessary example datasets are included in the container.
The RSV assembly example from the tour will take about 1.5 hour to run on four cores of a modern
CPU. If you are running it on a laptop or other personal workstation, you might want to check
your power saving setting in the OS to make sure the system will not go to sleep when you are not
interacting with it. For example, on the Mac you should check the box
Prevent computer from sleeping automatically when the display is off.
The Quick Start command above will use transient anonymous Docker volumes for both temporary working files during pipeline execution and the final outputs stored as Galaxy datasets. This has several implications:
- Once you stop the container (
docker stop) or reboot the host, your output data and all Galaxy histories will disappear - The file system where your Docker installation creates volumes should have enough free space to hold the temporary pipeline data. The pipeline caches all intermediate output files during its run for the ability to automatically retry any failed workflow steps. This means that it will use a total of 10 GB at peak time in the case of the eight RSV genomes example running on four CPU cores. Higher degree of parallelism (more CPU cores in the container) will increase peak disk use because more transient work files will coexist in the workflow at any given time. On Mac, Docker volumes typically reside on the same file system as the host operating system (OS). On Linux, the volumes might be on a relatively small separate partition that can overfill easily.
For the use beyond running the Quick Start example, you might want to supply a "bind-mount" that will make your outputs persist across container restarts and allow using any location you want for the pipeline temporary data:
docker run --rm -d -p 8080:80 --name ngs-mstb \
-v "/host/directory/path":"/export" \
-e GALAXY_CONFIG_SINGLE_USER=admin_ge@ngs-mstb.nowhere ngsmstb/ngs-mstb:latest
Above, we used the Unix shell line continuation symbol \ to split the long
command across three lines. You should remove this symbol if concatenating
the command into a single line.
In the command above, the placeholder /host/directory/path should be replaced with the actual absolute path to the existing directory on your
host machine. That directory should be empty when you first run this command and be located on a file system with enough free space. Galaxy will stage multiple files into that directory when it first starts. Because of that copying process, it might take up to five minutes for the
Galaxy UI to become available in the Web browser.
If you restart the container and use the same command later, Galaxy will notice the existing files and restore your previous session including job histories and output datasets.
Note on space for the images: Your Docker installation will still need to have enough free space to keep the container image itself (about 6GB). If you try a lot of different containers, this space fills up quickly, which creates a common source of problems encountered while using Docker in general. The housekeeping commands
such as docker system prune and docker volume prune in case of Docker
might help in such situations. You can also use these commands to clean up all space used by Docker
after you are done with using the image for good and stopped the container with
docker stop ngs-mstb. Note: the prune command will remove all unused images and stopped containers, not just NGS-MSTB, so you should be careful with using it.
Note on file sharing from Docker VM: On hosts that use a VM under the hood to run Docker (MacOS),
you might have to additionally expose the location of the /host/directory/path
to the VM. For example, on Mac Docker Desktop this is done through
Preferencies->File Sharing menu.
Use docker stop ngs-mstb to stop the container. The image will still be present. If you need to
remove the image, you can use docker rmi ngsmstb/ngs-mstb:latest.
Galaxy datafiles in the host directory that you bind-mounted to /export will not be deleted when the
image is deleted. You will have to use OS commands to remove them. You will probably have to invoke
administrative priviliges in the host OS because the files under that
directory will be created by the user galaxy within the container rather than by your current user
account in the host OS. The alternative approach is to delete them using the container:
docker run --rm --user root \
-v "/host/directory/path":"/export" \
ngsmstb/ngs-mstb:latest rm -rf /export
MGS-MSTB needs two types of externally provided data files:
- FASTQ files with paired-end NGS reads provided as two separate files for forward and reverse reads per each sequencing sample.
- One multi-FASTA file with recruiting references. Depending on the the analysis objectives, the same file can be used as QC references for aligning assembly contigs, or a second multi-FASTA file can be supplied for this purpose.
The FASTQ files should be named in a consistent pattern to allow configuring
the regular expressions for extracting sample IDs and establishing correct pairing
between forward and reverse read files. The extracted sample IDs will be used throughout
the pipeline to name rows in various output tables and to name the assembly
contigs in the output FASTA files. An example of one file naming convention is
SMA1396654_S1_L001_R1_001.fastq.gz for the forward and
SMA1396654_S1_L001_R2_001.fastq.gz for the reverse reads where
SMA1396654 is the sample ID. The inline Help
in the NGS-MSTB Generate Manifest... Galaxy tool explains how to tune the tool
parameters in order to accomodate a wide variety of possible file naming patterns.
Let us assume that you have placed the FASTQ files under some directory named
/path/to/seqstore/reads on the host file system where you start your Docker container.
Replace /path/to/seqstore/reads everywhere in these instructions with your actual absolute path.
Then, you should bind-mount this directory under a specific location
/seqstore/data in the container.
docker run --rm -d -p 8080:80 --name ngs-mstb \
-v "/path/to/seqstore/reads":"/seqstore/data" \
-v "/host/directory/path":"/export" \
-e GALAXY_CONFIG_SINGLE_USER=admin_ge@ngs-mstb.nowhere ngsmstb/ngs-mstb:latest
While you should always use this fixed /seqstore/data location after the :
in the bind-mount argument, the actual location of your files on the host OS
(the string before the :) can be any path.
The net result of setting this Docker parameter is that the NGS-MSTB software running
inside the container will see all files from /path/to/seqstore/reads directory on the
host machine to appear under /seqstore/data directory in the container.
The container will only need read-only access to the FASTQ files. Note:
the pipeline in the container will be executing as user galaxy that
has user ID (UID) 1450 and the same group ID (GID). You should either
create a user/group with the same UID/GID and give read and directory
execute permissions to either user or group to all files under your
/path/to/seqstore/reads, or make /path/to/seqstore/reads readable by all
users (if that is admissible for you security-wise). The latter can be
done with the following command on Linux or MacOS:
chmod -R o+rX "/path/to/seqstore/reads".
Note: Check this if you are running Docker on MacOS.
Pairs of FASTQ files across multiple samples can be spread across several subdirectories
under /path/to/seqstore/reads - please see the inline Help of the NGS-MSTB
Generate Manifest... Galaxy tool. The Help describes both
multi-directory search patterns and the regular expressions for extracting
the sample IDs.
When specifying path to the data subdirectory in the NGS-MSTB Generate Manifest... Galaxy
tool, the path should be given as relative to the fixed root path /seqstore inside the container.
This is easier to demonstrate through this key example:
Let us assume that on your host machine a batch of FASTQ files that you want to assemble
is located under a subdirectory /path/to/seqstore/reads/my_sequencing_run1, and their names end
in .fastq.gz. You bind-mounted your entire read store with a Docker parameter
-v "/path/to/seqstore/reads":"/seqstore/data". Then, in the Generate Manifest...
tool, you should supply: data/my_sequencing_run1/*.fastq.gz. Notice that the latter path is
a relative one - it starts with data rather than /data.
NGS-MSTB restricts the allowed file paths to be relative to the /seqstore root
as a security precaution in order to prevent the Web users from accessing arbitrary paths
inside the running container. The built-in example FASTQ files in the container are located under
/seqstore/test_data so that they would not clash with your own files bind-mounted
under /seqstore/data.
You might be running Docker container on a remote machine where you do not have your
sequencing inputs already on a file system that you could bind-mount into the container.
In that case, you can upload your FASTQ files into the container using any SFTP client.
The command below exposes a port 8022 on the host for connecting the remote SFTP client.
The SFTP server
already runs inside the container. Note that we still bind-mount a directory to
persist Galaxy datasets on the host. The SFTP server uses this location
to store the uploaded files (under a subdirectory ftp). If you change the port in the
docker run command, you need to change it accordingly when you access the SFTP server
from your client. We assume in this example that the DNS name of your remote host is my-host.
You should replace it with the actual hostname.
docker run --rm -d -p 8080:80 -p 8022:22 --name ngs-mstb \
-v "/host/directory/path":"/export" \
-e GALAXY_CONFIG_SINGLE_USER=admin_ge@ngs-mstb.nowhere ngsmstb/ngs-mstb:latest
To upload files, you can use any SFTP client such as Cyberduck,
FileZilla or a command-line sftp and connect to
your host my-host.
Important: You should use the full Galaxy user email as the SFTP user name, including
the components after the @ symbol, for example, admin_ge@ngs-mstb.nowhere. Use the
same password that you used to log that user into Galaxy.
Specify the custom port 8022 in the connection dialog to override the default SFTP port
that is 22.
Example sftp command: sftp -P 8022 admin_ge@ngs-mstb.nowhere@my-host.
Let us suppose that you uploaded your FASTQ files into a subdirectory my_sequencing_run1
in the SFTP server, and their names end in .fastq.gz. Then, in the Generate Manifest...
Galaxy tool, you should supply: ftp/my_sequencing_run1/*.fastq.gz.
If you, instead, placed the same files
directly under the SFTP server root directory outside of any subdirectories, you would
use in Galaxy ftp/*.fastq.gz. Note: Inside the generated manifest, the Galaxy user
email will get automatically inserted into the file paths like this
ftp/admin_ge@ngs-mstb.nowhere/SMA1396654_S1_L001_R1_001.fastq.gz. This is because internally,
the SFTP server is configured to keep each user's files in separate spaces.
These should be uploaded into your Galaxy history through the Web browser using Galaxy Get Data tool. Please see the interactive tour in the NGS-MSTB Galaxy container.
In the commands above, the environment variable -e GALAXY_CONFIG_SINGLE_USER=admin_ge@ngs-mstb.nowhere instructs Galaxy to starts in a single-user mode and automatically login the default Galaxy administrator account. Although you can use Galaxy UI in an anonymous session without logging in, you will not get any persistence of your work history if your restart the container or even close the browser window. This is why we used the automatic login above. However, this is only acceptable if your Galaxy container is executed on an isolated host and not shared between several users. If you want to use it in a shared environment, you should at least protect the Galaxy admin access by overriding the default values
of several access keys:
docker run --rm -d -p 8080:80 --name ngs-mstb -v "/host/directory/path":"/export" \
-e GALAXY_CONFIG_MASTER_API_KEY=your_unique_master_key \
-e GALAXY_DEFAULT_ADMIN_PASSWORD=your_unique_admin_password \
-e GALAXY_DEFAULT_ADMIN_KEY=your_unique_admin_key \
-e NGS_MSTB_SIG_KEY=your_unique_signature_key \
-e NGS_MSTB_GALAXY_REPORTS_PASSWORD=your_unique_galaxy_reports_password \
ngsmstb/ngs-mstb:latest
Replace each your_unique_* placeholder above with some secret string. The default
values of all those variables baked into the Docker image can be found inside the
Dockerfile in the ngs-mstb-docker repository or printed by inspecting the image
with docker inspect ngsmstb/ngs-mstb:latest. In particular, default admin user
is admin_ge@ngs-mstb.nowhere with a default password NgsMstb20.
You can login as the admin user and access the standard Galaxy Admin menu to create additional users for a shared deployment scenario.
You can look at the section "How to run - Docker for a validated GCP deployment" for more complete set of options of configuring production secure deployments of NGS-MSTB.
As part of ngs-mstb-docker repository, we have provided the deployment
scripts which pull the key configuration parameters from another repository
called ngs-mstb-secrets. The latter is just the example. The overall design
assumes that you would create a private copy of the ngs-mstb-secrets, tag it and
populate it with your own secret values for the critical access keys, Galaxy
Web page branding, certificates for HTTPs encrypted communication and
LDAP authentication config. These can be customized inside the same repository to configure several target hosts
on which you would want to deploy the NGS-MSTB container. That allows easily
deploying on the development, testing and production VMs as it is typical in
a validated software environment.
Under the ngs-mstb-docker repository, we have provided an example
set of instructions in a file example_prepare_host.sh. These instructions demonstrate how to prepare the Linux host
running Red Hat Enterprise Linux for deploying the NGS-MSTB container.
The instructions include creating the matching accounts in the host OS
and setting proper file permissions on the data directories.
Once the host was prepared, the script docker_deploy.sh can be executed.
This script will pull the customized configs and other run-time artifacts
from the ngs-mstb-secrets for the target host and launch the container
using the Docker Compose service definition. The latter will ensure that
the container will get automatically restarted with the same configuration
every time the host is rebooted.
Note on importing histories: Our support for GCP includes built-in checks that the intermediate
Galaxy datasets such as the manifest and the ARIBA reference pack have not been
modified by the analyst (through download, editing and re-upload sequence of
steps).
These checks are active in the tools all the time, even in non-GCP
deployments. The current implementatiom has a side effect that
if the user imports an existing Galaxy history,
the Extract target genes... tool will refuse to run with the manifest and
the ARIBA pack that were already present in the imported history. Both will have to
be rebuilt. The tools which build these datasets complete within seconds.
You can start with the interactive tours linked from the front page of the NGS-MSTB container UI. As the next step, the NGS-MSTB Galaxy tools provide extensive Help strings linked to all parameters, as well as tool-level Help sections located below the Execute button on each tool form. Those tool-level Help sections describe what the tool does, its place in the overall workflow and the outputs. Each tab of the main NGS-MSTB Web report starts with a short description of its content.
The NGS-MSTB Docker image is built on top of the Galaxy Docker image. The front page of that upstream repository describes in detail numerous additional configuration options which can be applied to further modify the behaviour of our derived image.
The software includes a testing suite that is composed from both unit tests and integration tests. The integration tests cover assembling various common and corner cases and comparing against the expected outputs. They also include checks for determinism. Those are implemented by running the same set of samples in multiple replicates and in several batches, and then comparing outputs for complete identity across replicates and batches.
The anonymized input sequencing reads used in the tests are available from
Zenodo NGS-MSTB community page.
In the repository ngs-mstb-docker, the script docker_run_tests.sh will execute the tests
inside a container.