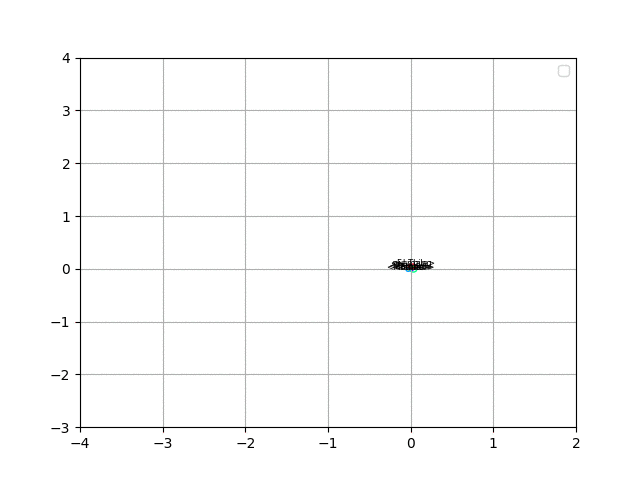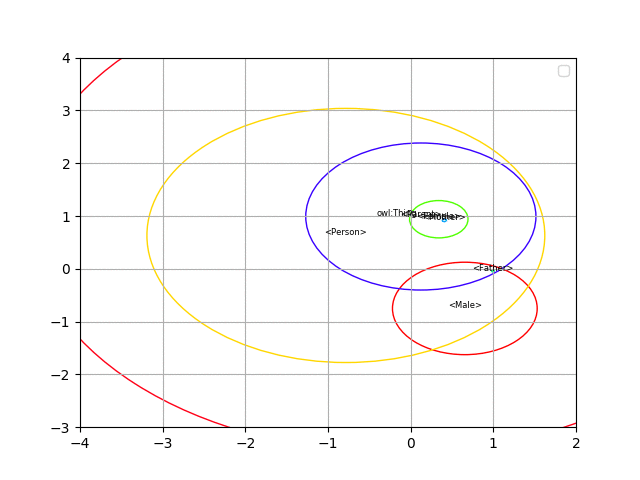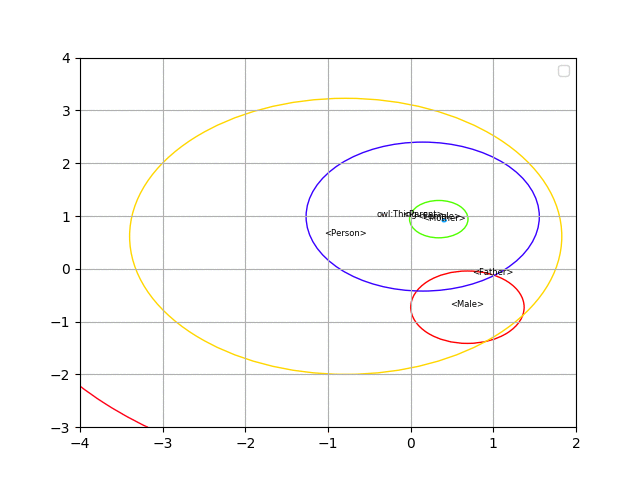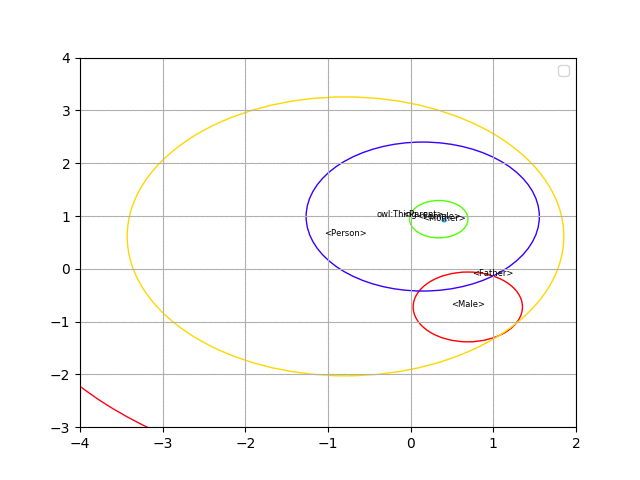
Here you can find the code to generate spherical embeddings for the description logic EL++ (the logic used in OWL 2 EL).
There are two parts that need to be executed sequentially. The first part converts an ontology in OWL 2 EL into a set of one of four normal forms. The second part of the method generates the embeddings for classes, relations, and individuals.
To build: To generate the normal forms, you need a modified version of the jcel reasoner which you find in a submodule here (if you have not cloned this repository with submodules, try
to do git clone --recurse-submodules https://github.com/bio-ontology-research-group/el-embeddings.
Next, cd jcel and mvn install to build the jcel jar files.
For convenience, we include the jar file in the jar/ subdirectory, so you may simply want to add jcel.jar to your CLASSPATH.
Then run groovy Normalizer.groovy and follow the instructions (i.e., command line options are in input OWL file and and output file containing the normal forms in OWL functional syntax).
usage: groovy Normalizer.groovy -i INPUT -o OUTPUT [-h]
-h,--help this information
-i,--input <arg> input OWL file
-o,--output <arg> output file containing normalized axioms
To generate the embeddings, run python elembedding.py --help and
follow instructions. You need CUDA installed to use a GPU, and need to
install python libraries with:
pip install -r requirements.txt
Usage: python elembedding.py [OPTIONS]
Options:
-df, --data-file TEXT Normalized ontology file (Normalizer.groovy)
-vdf, --valid-data-file TEXT Validation data set
-ocf, --out-classes-file TEXT Pandas pkl file with class embeddings
-orf, --out-relations-file TEXT
Pandas pkl file with relation embeddings
-bs, --batch-size INTEGER Batch size
-e, --epochs INTEGER Training epochs
-d, --device TEXT GPU Device ID
-es, --embedding-size INTEGER Embeddings size
-rn, --reg-norm INTEGER Regularization norm
-m, --margin FLOAT Loss margin
-lr, --learning-rate FLOAT Learning rate
-lhf, --loss-history-file TEXT Pandas pkl file with loss history
--help
Data is available here https://bio2vec.cbrc.kaust.edu.sa/data/elembeddings/el-embeddings-data.zip
Embeddings for yeast and human https://bio2vec.cbrc.kaust.edu.sa/data/elembeddings/embeddings.tar.gz
If you use El-Embeddings for your research, or incorporate our learning algorithms in your work, please cite:
Maxat Kulmanov, Wang Liu-Wei, Yuan Yan and Robert Hoehndorf; EL Embeddings: Geometric construction of models for the Description Logic EL++, arXiv:1902.10499