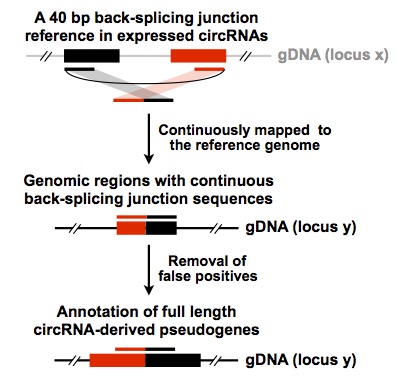
CIRCpseudo is a pipeline to map back-splicing junction sequences for circRNA-derived pseudogenes detection. Using this pipeline, you could previously detect candidate circRNA-derived pseudogenes in genome, after maual check you could do further characterizations for them.
- Not specific to certain species
- Aim to circRNA-derived pseudogenes detection
Required:
--circ CircRNA file (CIRCexplorer format file)
--ref Reference annotation file (refFlat format file)
--genome Reference genome file (Fasta format file)
--bwaidx Bwa index of reference genome
--output Output file
Optional:
--mismatch max mismaches between fusion sequences and genome, defalt 4
--fusionlen fusion lenth of back-splice exon-exon junctions defalt 40*Please add the CIRCpseudo directory to your $PATH first.
CIRCpseudo.pl -circ circRNA.bed -ref mm10_ref.txt -genome mm10.fa -bwaidx index/mm10.fa.idx -output mouse_pseudo.txt- ref.txt is in the format (Gene Predictions and RefSeq Genes with Gene Names) below (see details in the example file)
| Field | Description |
|---|---|
| geneName | Name of gene |
| isoformName | Name of isoform |
| chrom | Reference sequence |
| strand | + or - for strand |
| txStart | Transcription start position |
| txEnd | Transcription end position |
| cdsStart | Coding region start |
| cdsEnd | Coding region end |
| exonCount | Number of exons |
| exonStarts | Exon start positions |
| exonEnds | Exon end positions |
- mm10.fa is genome sequence in FASTA format.
You should get result file by --output. Output file will report Host gene location, Host gene name, Fusion sequence, Pseudogene location and Mismatches.
- CircRNA file in CIRCexplorer format
- Perl v5.14.1
- Bowtie v0.12.9
- BWA v0.6.2
- bedtools v2.19.0
Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL and Yang L. CircRNA-derived pseudogenes. Cell Res, 2016
Copyright (C) 2016 YangLab. See the LICENSE file for license rights and limitations (MIT).