MOGDx
Introduction
Multi-omic Graph Diagnosis (MOGDx) is a tool for the integration of omic data and classification of heterogeneous diseases. MOGDx exploits a patient similarity network framework to integrate omic data using Similarity Network Fusion (SNF) 1. One autoencoder per omic modality is trained and the latent embeddings from each autoencoder are concatenated. These reduced vectors are used as node features in the integrated network. Classification is performed on the fused network using the Graph Convolutional Network (GCN) deep learning algorithm 2. GCN is a novel paradigm for learning from both network structure and node features. Heterogeneity in diseases confounds clinical trials, treatments, genetic association and more. Accurate stratification of these diseases is therefore critical to optimize treatment strategies for patients with heterogeneous diseases. Previous research has shown that accurate classification of heterogenous diseases has been achieved by integrating and classifying multi-omic data 345. MOGDx improves upon this research. The advantages of MOGDx is that it can handle both a variable number of modalities and missing patient data in one or more modalities. Performance of MOGDx was benchmarked on the BRCA TCGA dataset with competitive performance compared to its counterparts. In summary, MOGDx combines patient similarity network integration with graph neural network learning for accurate disease classification.
Workflow
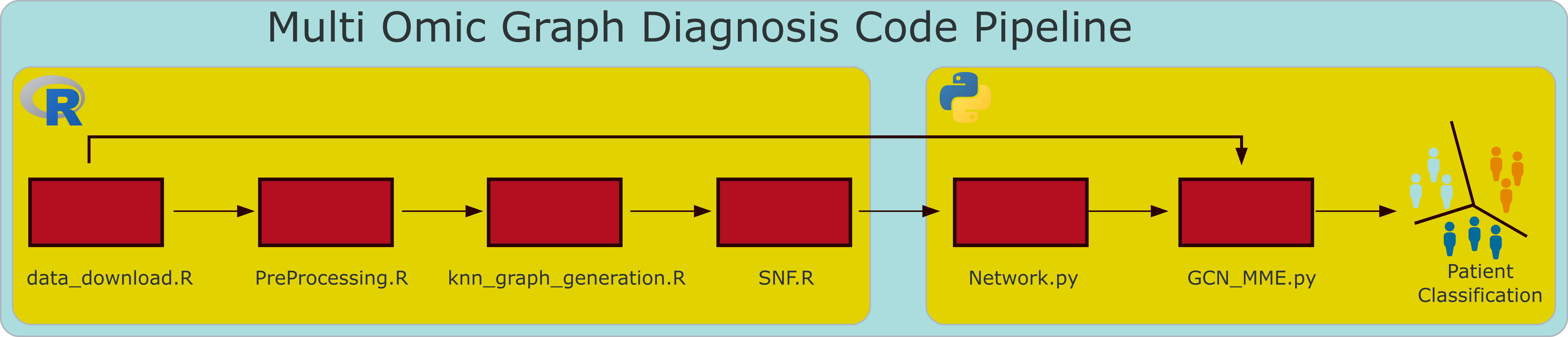
Full pipeline overview
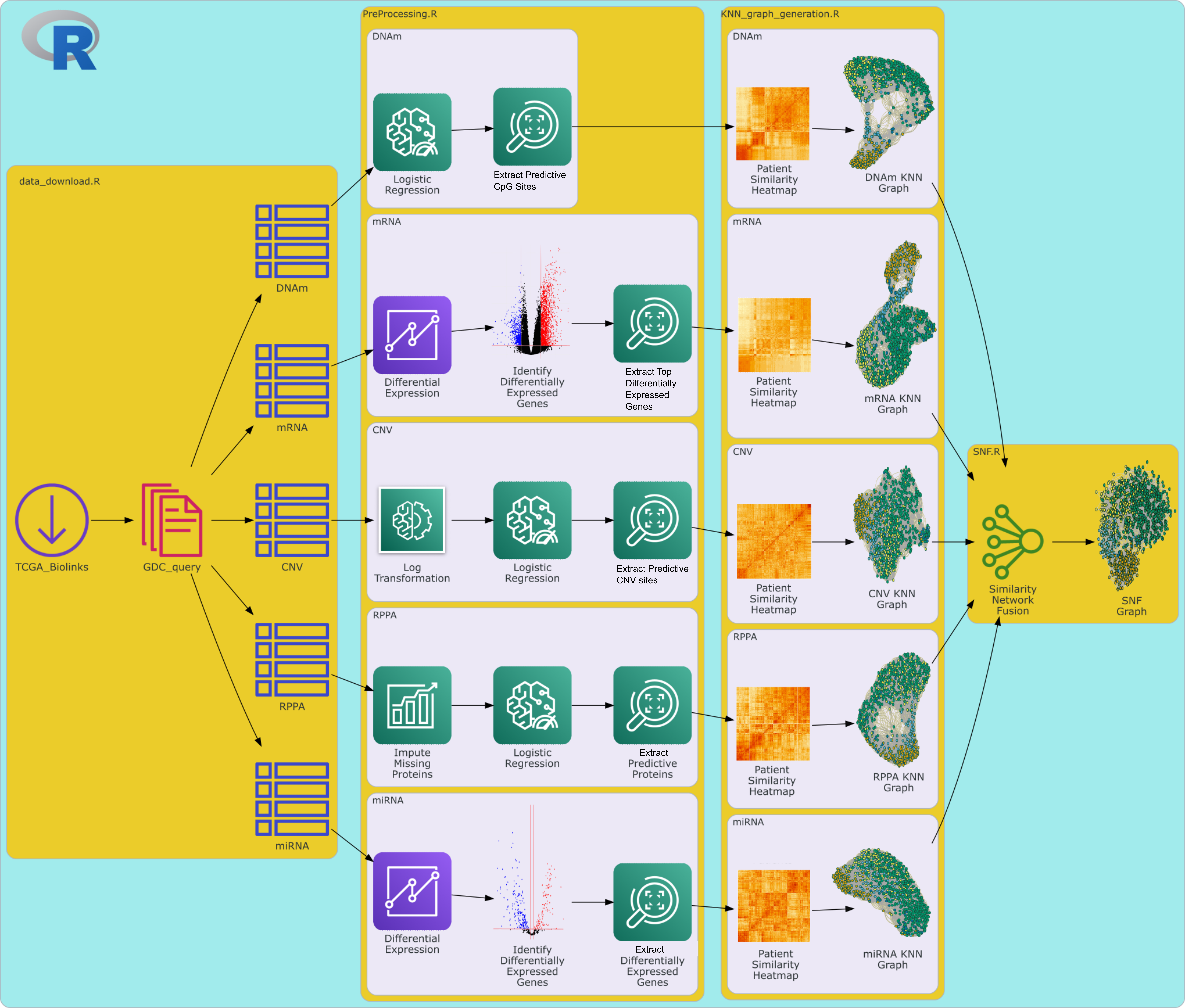
Pre-processing and Graph Generation
AE, GNN and Heterogenous Disease Classification
Installation
A working version of R and Python is required. R version 4.2.2 and Python version 3.9.2 was used to obtain all results.
Steps 1-4 are executed in R, while step 5 is designed to run on a cluster with a GPU but can be executed locally if there is sufficient memory availability.
Step 1 - Data Download
Use the R script data_download.R to download all data changing the project to BRCA/LGG/KICH/KIRC/KICH
Step 2 - Preprocessing
Create a folder called data and a folder for each dataset with naming convention 'TCGA-' e.g. 'TCGA-BRCA' inside this folder.
Run the R script Preprocessing.R specifying the phenotypical trait and project.
The options are
BRCA :
project = 'BRCA'
trait = 'paper_paper_BRCA_Subtype_PAM50'
LGG :
project = 'LGG'
trait = 'paper_Grade'
Note : To create the KIPAN dataset, the KIRC, KICP and KICH datasets have to be combined. This can be achieved by copying the downloaded files from all three seperate datasets into a single dataset called KIPAN keeping the same naming structures. A column named 'subtype' specifying which dataset the patient came from needs to be created in the Meta data file. A basic knowledge of R is required for this.
KIPAN :
project = 'KIPAN'
trait = 'subtype'
Step 3 - Graph Generation
Create a folder called Network, to store all graphs. Within this folder create a folder for each modality. Each modalities graph will be saved inside their respective folder with name graph.csv.
Use the R script knn_graph_generation.R specifying the phenotypical trait, project and modalities downloaded in the for loop.
Step 4 - SNF
Create a folder in Network called SNF
Copy each modalities graph.csv to this folder with naming convention 'modality_graph.csv' e.g. 'mRNA_graph.csv' \
Specify the modalities of interest in the list mod_list
Run the R script SNF.R
Step 5 - Execute MOGDx.py
Copy all expression and meta files for all modalities into a single folder with naming convention 'modality_datExpr.csv'/'modality_datMeta.csv' e.g. 'mRNA_datExpr.csv'/'mRNA_datMeta.csv'.
These files will have been created in the directory ./data/TCGA-BRCA/mRNA/datExpr.csv for the mRNA expression file in the BRCA project
Copy the patient similarity network in ./Network/SNF/graph.csv to this folder also
MOGDx is a command line tool. A sample command is :
python MOGDx.py -i "/raw_data/raw_BRCA" -o "./Output/BRCA/" -snf "graph.csv" --n-splits 5 -ld 32 64 64 32 32 --target "paper_BRCA_Subtype_PAM50" --index-col "patient" --epochs 2500 --lr 0.01 --layer-activation "elu" "elu" --layers 128 128
-i is the location of the raw data containing all datExpr, datMeta and graph.csv files
-o is the location where the output will be printed
-snf is the name of the fused psn
--n-splits is the number of cross validation splits
-ld is the latent dimension per modality. It is order alphabetically
--target is the phenotype being classified
--index-col is the column containing the patient ids
All other arguments are related to the Graph Convolutional Network model
Requirements
All requirements are specified in requirements.txt
To create virtual env execute :
conda create --name <env> --file requirements.txt
Contact
Barry Ryan
PhD Candidate Biomedical AI, University of Edinburgh
School of Informatics, 10 Crichton Street
Email: barry.ryan@ed.ac.uk
LinkedIn: https://www.linkedin.com/in/barry-ryan/
Citations
Footnotes
-
Bo Wang et al. “Similarity network fusion for aggregating data types on a genomic scale”. en. In: Nature Methods 11.3 (Mar. 2014). Number: 3 Publisher: Nature Publishing Group, pp. 333–337. ISSN: 1548-7105. DOI: 10.1038/nmeth.2810. URL: https://www.nature.com/articles/nmeth.2810 (visited on 11/07/2022) ↩
-
Thomas N. Kipf and Max Welling. Semi-Supervised Classification with Graph Convolutional Networks. en. arXiv:1609.02907 [cs, stat]. Feb. 2017. URL:http://arxiv.org/abs/1609.02907 (visited on 09/26/2022). ↩
-
Shraddha Pai et al. “netDx: interpretable patient classification using integrated patient similarity networks”. In: Molecular Systems Biology 15.3 (Mar. 2019). Publisher: John Wiley & Sons, Ltd, e8497. ISSN: 1744-4292. DOI: 10.15252/msb.20188497. URL: https://www.embopress.org/doi/full/10.15252/msb. 20188497 (visited on 12/05/2022). ↩
-
Xiao Li et al. “MoGCN: A Multi-Omics Integration Method Based on Graph Convolutional Network for Cancer Subtype Analysis”. eng. In: Frontiers in Genetics 13 (2022), p. 806842. ISSN: 1664-8021. DOI: 10.3389/fgene.2022.806842. ↩
-
Tongxin Wang et al. “MOGONET integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification”. en. In: Nature Communications 12.1 (June 2021). Number: 1 Publisher: Nature Publishing Group, p. 3445. ISSN: 2041-1723. DOI: 10.1038/s41467-021-23774-w. URL: https://www.nature.com/articles/s41467-021-23774-w (visited on 01/26/2023). ↩