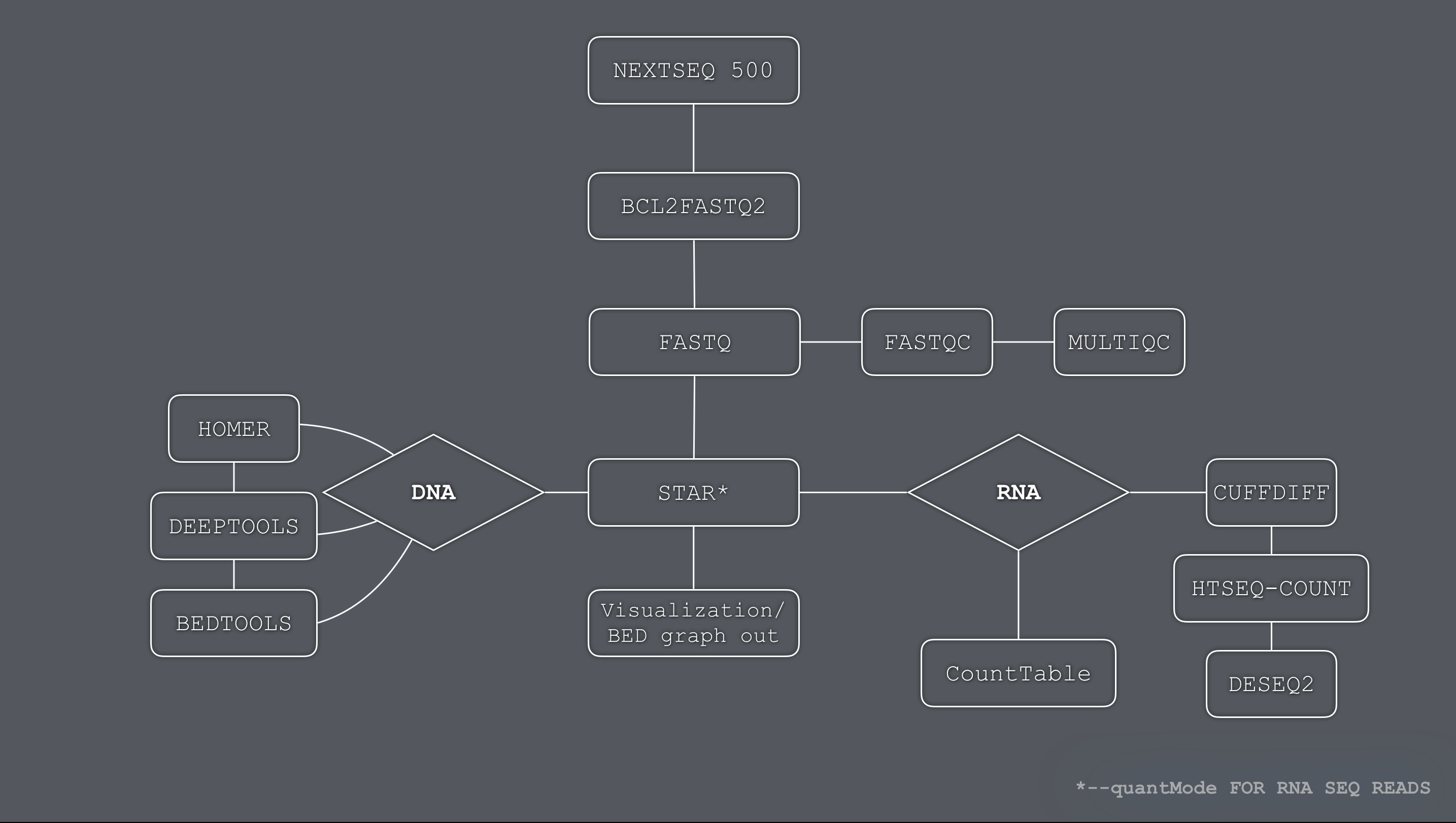
bcl2fastq.HPC.sh:
Demultiplexes Illumina NextSeq 500 samples (bcl intensities to fastq).
Performs quality control assessments using FASTQC.
Generates a single html report plotting all quality control metrics.
Usage:
$ sbatch bcl2fastq.HPC.sh NameOfRunFolder NameOfSampleSheet NameOfPI
barcodeSplit.pl:
Demultiplexes fastq reads based on barcodes of interest.
reHash.py:
Rename utility.
reveRse.py:
Generates i5 reverse complements for Illumina NextSeq 500 samples.
Usage:
$ python reveRse.py < indices.txt >
star.align.HPC.sh:
Aligns PE or SE fastq samples to reference genome using star ultrafast aligner.
Generates bedgraph files for UCSC genome browser visualization.
Usage:
$ sbatch star.align.HPC.sh <SE or PE> <ORG> <ASSEMBLY>
DESeq2.R:
Performs differential gene expression analysis of RNA-seq samples.
(rawCounts used are either from STAR --quantMode GeneCounts or HTSEQ outputs)
Usage:
$ RScript DESeq2.R <Experiment.Name> <Numerator> <Denominator>
For DESeq2 processCounts.sh and countMatrix.sh helper scripts are available for
preprocessing raw counts and creating the countMatrix.txt file, needed by the DESeq2.R script.
Usage:
$ processCounts.sh <1> or <2>