slendr simulation workshop at MPI EVA (2023)
You can find the slides here.
Here is a render of the slides as a single HTML page (easier for reference).
This README describes how to set up your machine for the practical workshop. After you're done installing everything, make sure to run a small testing simulation (here) to make sure that everything works as needed.
- Working R installation(at least R 3.6, preferably 4.x)
- RStudio highly recommended
- macOS / Linux / Windows (the latter won't support SLiM-based simulations, but that's OK!)
Getting slendr to work is critical. The whole workshop is dedicated to this package.
You can install slendr with this:
install.packages("slendr")
After installation, load slendr itself.
library(slendr)
This will very likely write a message that:
-
you are missing SLiM -- this is OK, feel free to ignore this
-
you are missing a Python environment -- we will fix this in the next step.
Next, run the following command. This will ask for permission to install an isolated Python mini-environment just for slendr -- this won't affect your own Python setup at all, so don't be afraid to confirm this!
setup_env()
Finally, make sure you get a positive confirmation from the following check:
check_env()
On future occasions, you will have to call init_env() after running library(slendr)!
Copy the following script to your R session after you successfully installed your R dependencies as described above.
First run this:
library(slendr)
init_env()
Followed by this:
o <- population("outgroup", time = 1, N = 100)
b <- population("b", parent = o, time = 500, N = 100)
c <- population("c", parent = b, time = 1000, N = 100)
x1 <- population("x1", parent = c, time = 2000, N = 100)
x2 <- population("x2", parent = c, time = 2000, N = 100)
a <- population("a", parent = b, time = 1500, N = 100)
gf <- gene_flow(from = b, to = x1, start = 2100, end = 2150, rate = 0.1)
model <- compile_model(
populations = list(a, b, x1, x2, c, o), gene_flow = gf,
generation_time = 1, simulation_length = 2200
)
ts <- msprime(model, sequence_length = 1e6, recombination_rate = 1e-8)
ts_samples(ts)
If this runs without error and you get a small summary table with simulated samples, you're all set!
I will use some tidyverse packages for analysis and plotting.
I recommend you install the following packages:
install.packages(c("dplyr", "ggplot2", "cowplot", "magrittr"))
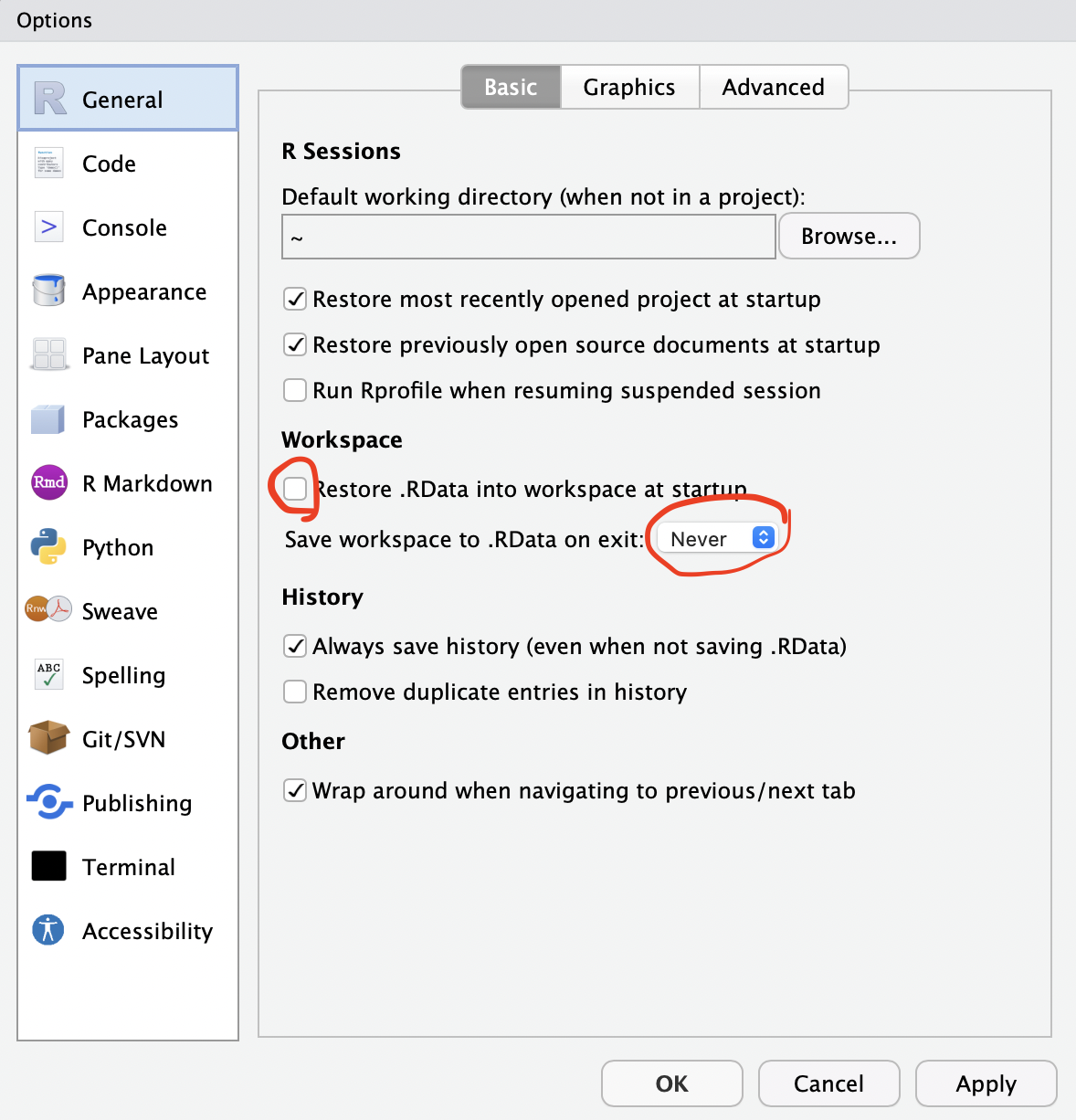
RStudio sometimes interferes with Python setup needed for simulation. To fix this, go to Tools -> Global Options in RStudio and set the following options:
This work is licensed under a Creative Commons Attribution 4.0 International License.