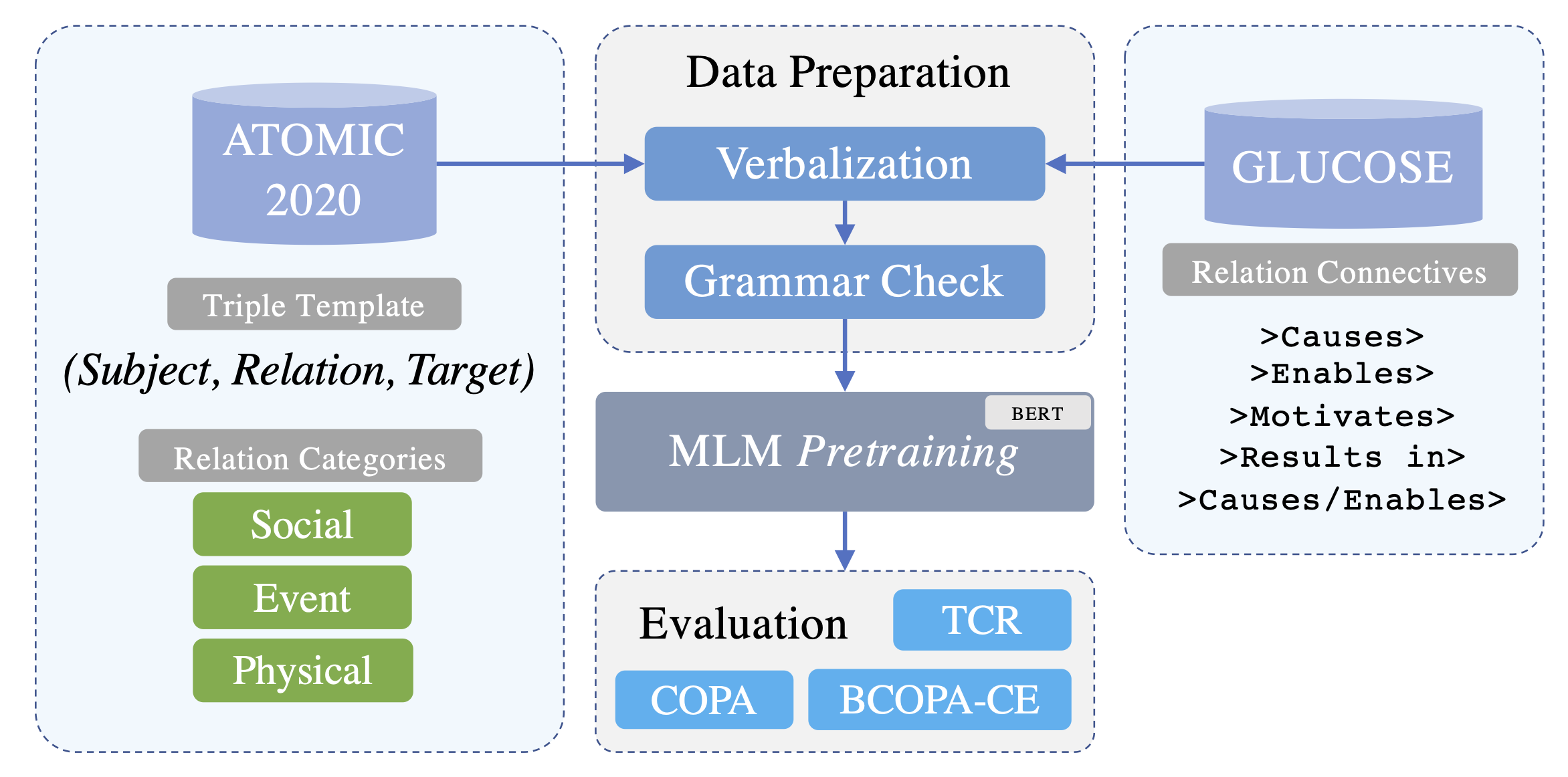
Triples in ATOMIC2020 are stored in form of: (subject, relation, target). We convert (verbalize) these triples to natural language text to later use them in training/fine-tuning some Pretrained Language Models (PLMs):
- Download ATOMIC2020 here, put the zip file in the
/datafolder, and unzip it (we only needdev.tsvandtrain.tsv). - Run the following code:
atomic_to_text.py(depending on whether you're running the grammar check, this may take a while.) - Outputs will be stored as
.txtand.csvfiles in the/datafolder following the name patterns:atomic2020_dev.*andatomic2020_train.*.
- Download GLUCOSE here, unzip the file, and put the
GLUCOSE_training_data_final.csvfile in the/datafolder. - Run the following code: glucose_to_text.py
- Output will be stored in:
data/glucose_train.csv
Once we verbalized the ATOMIC2020 knowledge graph and GLUCOSE dataset to text, we continually pretrain a Pretrained Language Model (PLM), BERT here, using the converted texts. We call this pretraining step a continual pretraining since we use one of the objectives, Masked Language Modeling (MLM) that was originally used in pretraining BERT, to further train the PLM. There are two steps for running the pretraining:
- Setting the parameters in the
config/pretraining_config.json: Even though most of these parameters are self-descriptive, we give a brief explanation about some of them for clarification purposes:relation_category(for ATOMIC2020): A list of triple types (strings) with which we want to continually pretrain our model. There are three main categories of triples in ATOMIC2020:event,social, andphysical. These categories may deal with different types of knowledge. And, models pretrained with each of these categories or a combination of them may give us different results when fine-tuned and tested on downstream tasks. As a result, we added an option for choosing the triple type(s) with which we want to run the pretraining.
- Runnig the pretraining code:
pretraining.py
Our models are deployed on the HuggingFace's model hub. Here is an example of how you can load the models:
from transformers import AutoTokenizer, AutoModel
# bert model examples
tokenizer_bert = AutoTokenizer.from_pretrained("bert-large-cased")
atomic_bert_model = AutoModel.from_pretrained("phosseini/atomic-bert-large")
glucose_model = AutoModel.from_pretrained("phosseini/glucose-bert-large")
# roberta model example
tokenizer_roberta = AutoTokenizer.from_pretrained("roberta-large")
atomic_roberta_model = AutoModel.from_pretrained("phosseini/atomic-roberta-large")Full list of models on HuggingFace
| Model | Training Data |
|---|---|
phosseini/glucose-bert-large |
GLUCOSE |
phosseini/glucose-roberta-large |
GLUCOSE |
phosseini/atomic-bert-large |
ATOMIC2020 event relations |
phosseini/atomic-bert-large-full |
ATOMIC2020 event, social, physical relations |
phosseini/atomic-roberta-large |
ATOMIC2020 event relations |
phosseini/atomic-roberta-large-full |
ATOMIC2020 event, social, physical relations |
After pretraining the PLM with the new data, it is time to fine-tune and evaluate its performance on downstream tasks. So far, we have tested our models on two benchmarks including COPA and TCR. In the following, we explain the fine-tuning process for each of them.
- Run the
convert_copa.pyto generate all the required COPA-related data files (train/test) for fine-tuning. - Setting the parameters in
config/fine_tuning_config.json. Description of some parameters:tuning_backend: For choosing the hyperparameter tuning backend,rayoroptunahyperparameter_search: Whether to run hyperparameter search or not.1for running and0for not running, respectively.cross_validation: Whether running the cross-validation on the development set or not. If thehyperparameter_searchis0, this parameter will be ignored since if we do not want to fine-tune the model with a known set of hyperparameters there is no need to run cross-validation on the development set.tuning_*: All parameters related to hyperparameter tuning.tuning_learning_rate_do_rangeis set to1when we want to search learning rates within a range instead of a predefined list of learning rate values.tuning_learning_rate_startandtuning_learning_rate_endare to specify the start and end of such a range. Alternatively, we can set thetuning_learning_rate_do_rangeto0and learning rates for hyperparameter tuning will be selected from thetuning_learning_ratelist.
- Running the fine-tuning code:
fine_tuning_copa.py
Best hyperparameter values for different models:
| Model | Epochs | Batch Size | Learning Rate |
|---|---|---|---|
bert-large-cased |
4 | 8 | 3e-05 |
phosseini/atomic-bert-large |
4 | 4 | 2e-5 |
phosseini/atomic-roberta-large |
4 | 4 | 1e-5 |
- Convert the original TCR dataset to a standard format using CREST. Then convert the new files to a format required by R-BERT. Check the following notebook to see how: tcr_data_preparation.ipynb
- Now run R-BERT using the following command (we slightly modified R-BERT, please find our forked repository here):
! python3 main.py \
--do_train \
--do_eval \
--model_name_or_path phosseini/atomic-bert-large \
--tokenizer_name_or_path bert-large-cased \
--num_train_epochs 10 \
--train_batch_size 8 \
--seed 117 \
--save_steps 40 \
--max_seq_len 200
@inproceedings{hosseini-etal-2022-knowledge,
title = "Knowledge-Augmented Language Models for Cause-Effect Relation Classification",
author = "Hosseini, Pedram and Broniatowski, David A. and Diab, Mona",
booktitle = "Proceedings of the First Workshop on Commonsense Representation and Reasoning (CSRR 2022)",
month = may,
year = "2022",
address = "Dublin, Ireland",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/2022.csrr-1.6",
pages = "43--48",
}