A workflow for the statistical analysis and visualization of differentially methylated regions (DMRs) of CpG count matrices (Bismark cytosine reports) from the CpG_Me pipeline.
- Overview
- DMR Approach and Interpretation
- Installation
- The Design Matrix and Covariates
- Input
- Output
- Citation
- Publications
- Acknowledgements
The goal of DMRichR is to make the comprehensive statistical analysis of whole genome bisulfite sequencing (WGBS) data accessible to the larger epigenomics community, so that it no longer remains a niche methodology. Whether it be peripheral samples from a large-scale human epidemiological study or a select set of precious samples from model and non-model organisms, WGBS can provide novel insight into the epigenome and its role in the regulation of gene expression. Furthermore, the functions and workflow are written with the goal of bridging the gap for those transitioning from Illumina's Infinium assay technology (450K and EPIC arrays) by providing statistical analysis and visualization functions that present the data in a familiar format.
The overarching theme of DMRichR is the synthesis of popular Bioconductor R packages for the analysis of genomic data with the tidyverse philosophy of R programming. This allows for a streamlined and tidy approach for downstream data analysis and visualization. In addition to functioning as an R package, the central component of DMRichR is an executable script that is meant to be run as a single call from command line. While this is a non-traditional approach for R programming, it serves as a novel piece of software that simplifies the analysis process while also providing a backbone to build custom workflows on (in a manner similar to a traditional vignette).
DMRichR leverages the statistical algorithms from two popular R packages,dmrseq and bsseq, which enable the inference of differentially methylated regions (DMRs) from low coverage WGBS. In these smoothing based approaches, CpG sites with higher coverage are given a higher weight and used to infer the methylation level of neighboring CpGs with lower coverage. This approach favors a larger sample size over a deeper sequencing depth, and only requires between 1-5x coverage for each sample. By focusing on the differences in methylation levels between groups, rather than the absolute levels within a group, the methodologies utilized allow for a low coverage WGBS approach that assays ~10x more of the genome for only around ~2x the price of competing reduced representation methods (i.e. arrays and RRBS). In our experience, it is these unexplored regions of the genome that contain the most informative results for studies outside of the cancer research domain; however, these regions should also provide novel insight for cancer researchers as well. In order to facilitate an understanding of these DMRs and global methylation levels, DMRichR also works as a traditional R package with a number of downstream functions for statistical analysis and data visualization that can be viewed in the R folder.
A single command line call performs the following steps:
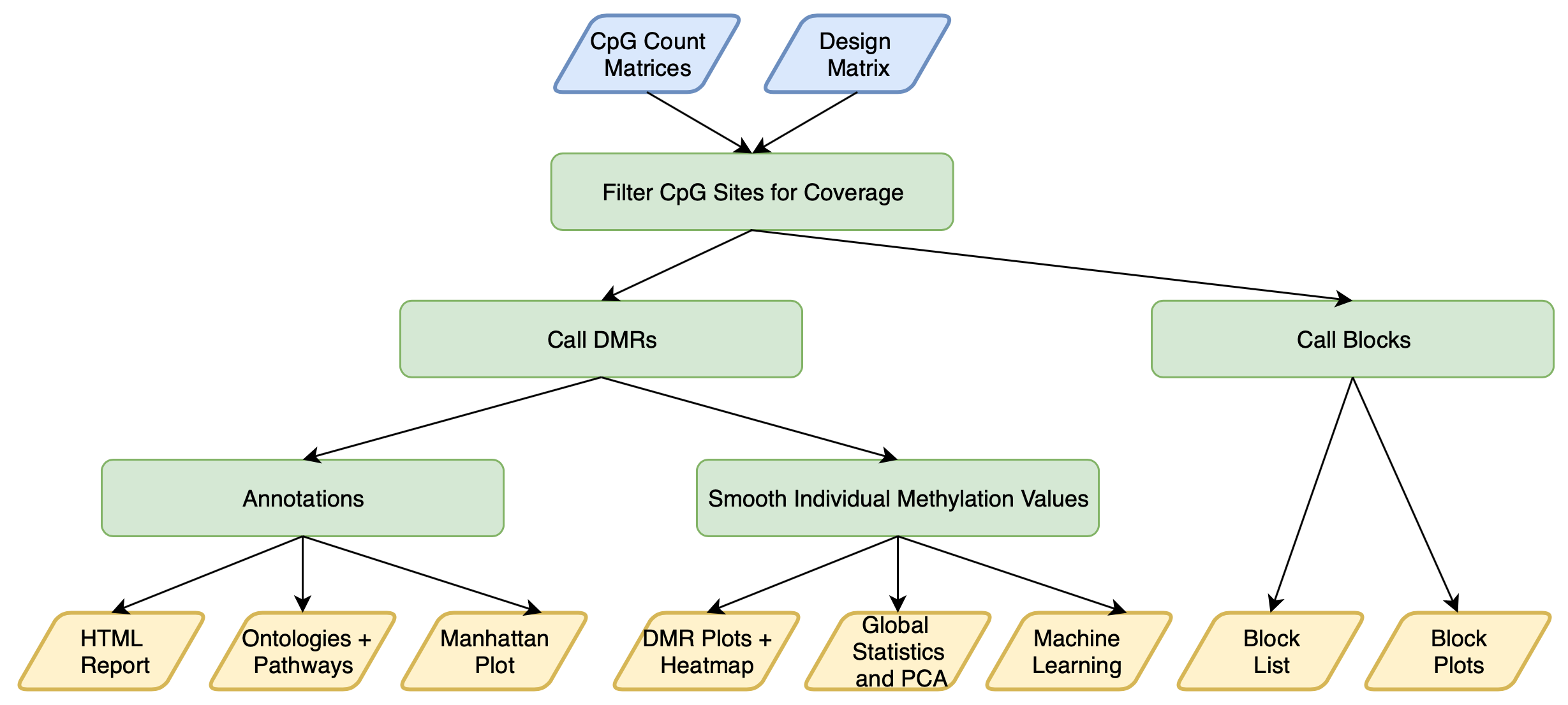
The main statistical approach applied by the executable script located in the exec folder is dmrseq::dmrseq(), which identifies DMRs in a two step approach:
- DMR Detection: The differences in CpG methylation for the effect of interest are pooled and smoothed to give CpG sites with higher coverage a higher weight, and candidate DMRs with a difference between groups are assembled.
- Statistical Analysis: A region statistic for each DMR, which is comparable across the genome, is estimated via the application of a generalized least squares (GLS) regression model with a nested autoregressive correlated error structure for the effect of interest. Then, permutation testing of a pooled null distribution enables the identification of significant DMRs. This approach accounts for both inter-individual and inter-CpG variability across the entire genome.
The main estimate of a difference in methylation between groups is not a fold change but rather a beta coefficient, which is representative of the average effect size; however, it is on the scale of the arcsine transformed differences and must be divided by π (3.14) to be similar to the mean methylation difference over a DMR, which is provided in the percentDifference column. Since the testing is permutation based, it provides empirical p-values as well as FDR corrected q-values.
One of the key differences between dmrseq and other DMR identification packages, like bsseq, is that dmrseq is performing statistical testing on the DMRs themselves rather than testing for differences in single CpGs that are then assembled into DMRs like bsseq::dmrFinder() does. This unique approach helps with controlling the false discovery rate and testing the correlated nature of CpG sites in a regulatory region, while also enabling complex experimental designs. However, since dmrseq::dmrseq() does not provide individual smoothed methylation values, bsseq::BSmooth() is utilized to generate individual smoothed methylation values from the DMRs. Therefore, while the DMRs themselves are adjusted for covariates, the individual smoothed methylation values for these DMRs are not adjusted for covariates.
You can also read my general summary of the drmseq approach on EpiGenie.
Example DMR
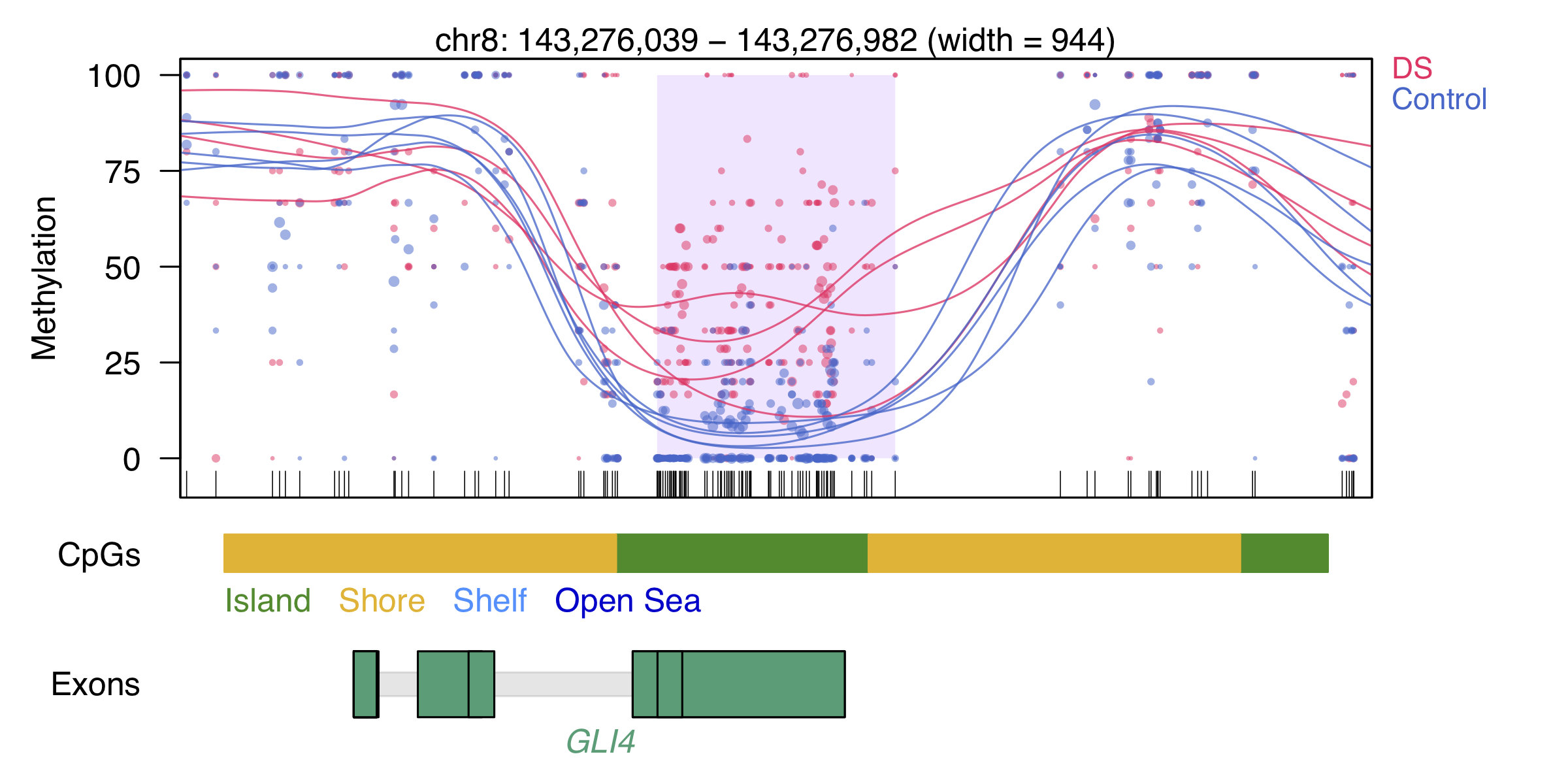 Each dot represents the methylation level of an individual CpG in a single sample, where the size of the dot is representative of coverage. The lines represent smoothed methylation levels for each sample, either control (blue) or DS (red). Genic and CpG annotations are shown below the plot.
Each dot represents the methylation level of an individual CpG in a single sample, where the size of the dot is representative of coverage. The lines represent smoothed methylation levels for each sample, either control (blue) or DS (red). Genic and CpG annotations are shown below the plot.
No manual installation of R packages is required, since the required packages and updates will occur automatically upon running the executable script located in the exec folder. However, the package does require Bioconductor, which you can install or update to using:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install(version = "3.10")
Additionally, if you are interested in creating your own workflow as opposed to using the executable script, you can download the package using:
BiocManager::install(c("remotes", "ben-laufer/DMRichR"))
This script requires a basic design matrix to identify the groups and covariates, which should be named sample_info.xlsx and contain header columns to identify the covariates. The first column of this file should be the sample names and have a header labelled as Name. In terms of the testCovariate label (i.e. Group or Diagnosis), it is important to have the label for the experimental samples start with a letter in the alphabet that comes after the one used for control samples in order to obtain results for experimental vs. control rather than control vs. experimental. You can select which specific samples to analyze from the working directory through the design matrix, where pattern matching of the sample name will only select bismark cytosine report files with a matching name before the first underscore, which also means that sample names should not contain underscores. Within the script, covariates can be selected for adjustment. There are two different ways to adjust for covariates: directly adjust values or balance permutations.
| Name | Diagnosis | Age | Sex |
|---|---|---|---|
| SRR3537014 | Idiopathic_ASD | 14 | M |
| SRR3536981 | Control | 42 | F |
DMRichR utilizes Bismark cytosine reports, which are genome-wide CpG methylation count matrices that contain all the CpGs in your genome of interest, including CpGs that were not covered in the experiment. The genome-wide cytosine reports contain important information for merging the top and bottom strand of symmetric CpG sites, which is not present in Bismark coverage and bedGraph files. In general, cytosine reports have the following pattern: *_bismark_bt2_pe.deduplicated.bismark.cov.gz.CpG_report.txt.gz. CpG_Me will generate a folder called cytosine_reports after calling the final QC script (please don't use the cytosine_reports_merged folder for DMRichR). If you didn't use CpG_Me, then you can use the coverage2cytosine module in Bismark to generate the cytosine reports. The cytosine reports have the following format:
| chromosome | position | strand | count methylated | count non-methylated | C-context | trinucleotide context |
|---|---|---|---|---|---|---|
| chr2 | 10470 | + | 1 | 0 | CG | CGA |
| chr2 | 10471 | - | 0 | 0 | CG | CGG |
| chr2 | 10477 | + | 0 | 1 | CG | CGA |
| chr2 | 10478 | - | 0 | 0 | CG | CGG |
Before running the executable, ensure you have the following project directory tree structure for the cytosine reports and design matrix:
├── Project
│ ├── cytosine_reports
│ │ ├── sample1_bismark_bt2.deduplicated.bismark.cov.gz.CpG_report.txt.gz
│ │ ├── sample2_bismark_bt2.deduplicated.bismark.cov.gz.CpG_report.txt.gz
│ │ ├── sample_info.xlsx
This workflow requires the following variables:
-g --genomeSelect either: hg38, mm10, rn6, or rheMac8.-x --coverageCpG coverage cutoff for all samples, 1x is the default and minimum value.-s --perGroupPercent of samples per a group for CpG coverage cutoff, values range from 0 to 1, 1 (100%) is the default.-m --minCpGsMinimum number of CpGs for a DMR, 5 is default.-p --maxPermsNumber of permutations for DMR and block analyses, 10 is default.-o --cutoffThe cutoff value for the single CpG coefficient utilized to discover testable background regions, values range from 0 to 1, 0.05 (5%) is the default.-t --testCovariateCovariate to test for significant differences between experimental and control, i.e. Diagnosis.-a --adjustCovariateAdjust covariates that are continuous or contain two or more factor groups, i.e. "Age". More than one covariate can be adjusted for using single brackets and the;delimiter, i.e.'Sex;Age'-m --matchCovariateCovariate to balance permutations, which is meant for two-group factor covariates in small sample sizes in order to prevent extremely unbalanced permutations. Only one two-group factor can be balanced, i.e. Sex. Note: This will not work for larger sample sizes (> 500,000 permutations) and is not needed for them as the odds of sampling an extremely unbalanced permutation for a covariate decreases with increasing sample size. Futhermore, we generally do not use this in our analyses, since we prefer to directly adjust for sex.-c --coresThe number of cores to use, 20 is recommended but you can go as low as 1, 8 is the default.
Below is an example of how to execute the main R script (DM.R) in the exec folder on command line. This should be called from the working directory that contains the cytosine reports.
call="Rscript \
--vanilla \
/share/lasallelab/programs/DMRichR/DM.R \
--genome hg38 \
--coverage 1 \
--perGroup '1' \
--minCpGs 5 \
--maxPerms 10 \
--cutoff '0.05' \
--testCovariate Diagnosis \
--adjustCovariate 'Sex;Age' \
--cores 20"
echo $call
eval $call
If you are using the Barbera cluster at UC Davis, the following commands can be used to execute DM.R from your login node (i.e. epigenerate), where htop should be called first to make sure the whole node is available. This should be called from the working directory that contains the cytosine reports and not from within a screen.
module load R/3.6.1
call="nohup \
Rscript \
--vanilla \
/share/lasallelab/programs/DMRichR/DM.R \
--genome hg38 \
--coverage 1 \
--perGroup '1' \
--minCpGs 5 \
--maxPerms 10 \
--cutoff '0.05' \
--testCovariate Diagnosis \
--adjustCovariate 'Sex;Age' \
--cores 60 \
> DMRichR.log 2>&1 &"
echo $call
eval $call
echo $! > save_pid.txt
You can then check on the job using tail -f DMRichR.log and ⌃ Control + c to exit the log view.
You can cancel the job from the project directory using cat save_pid.txt | xargs kill. You can also check your running jobs using ps -ef | grep , which should be followed by your username i.e. ps -ef | grep blaufer. Finally, if you still see leftover processes in htop, you can cancel all your processes using pkill -u, which should be followed by your username i.e. pkill -u blaufer.
Alternatively, the executable can also be submitted to the cluster using the shell script via sbatch DM.R.sh.
This workflow provides the following files:
- DMRs and testable background regions
- Individual smoothed methylation values for DMRs and background regions
- DMR plots of individual smoothed methylation values
- Smoothed global and chromosomal methylation values and statistics
- PCA plots of 20 Kb windows (all genomes) and CpG island windows (hg38, mm10, and rn6)
- Heatmap of individual smoothed methylation values for DMRs
- An HTML report of annotated DMRs
- Gene region and CpG annotations and plots (only for hg38, mm10, or rn6)
- Manhattan and Q-Q plots
- Gene ontology and pathway enrichment tables and plots (enrichr and GOfuncR for all genomes, GREAT for hg38 and mm10)
- Machine learning feature selection report and heatmap
- Large blocks of differential methylation and testable background blocks
- Block plots
If you use DMRichR in published research please cite the following 3 articles:
Laufer BI, Hwang H, Vogel Ciernia A, Mordaunt CE, LaSalle JM. Whole genome bisulfite sequencing of Down syndrome brain reveals regional DNA hypermethylation and novel disease insights. Epigenetics, 2019. doi: 10.1080/15592294.2019.1609867
Korthauer K, Chakraborty S, Benjamini Y, and Irizarry RA. Detection and accurate false discovery rate control of differentially methylated regions from whole genome bisulfite sequencing. Biostatistics, 2018. doi: 10.1093/biostatistics/kxy007
Hansen KD, Langmead B, Irizarry RA. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biology, 2012. doi: 10.1186/gb-2012-13-10-r83
The following publications utilize CpG_Me and DMRichR:
Wöste M, Leitão E, Laurentino S, Horsthemke B, Rahmann S, Schröder C. wg-blimp: an end-to-end analysis pipeline for whole genome bisulfite sequencing data. bioRxiv preprint. doi: 10.1101/859900
Mordaunt CE, Jianu JM, Laufer BI, Zhu Y, Dunaway KW, Bakulski KM, Feinberg JI, Volk HE, Lyall K, Croen LA, Newschaffer CJ, Ozonoff S, Hertz-Picciotto I, Fallin DM, Schmidt RJ, LaSalle JM. Cord blood DNA methylome in newborns later diagnosed with autism spectrum disorder reflects early dysregulation of neurodevelopmental and X-linked genes. bioRxiv preprint. doi: 10.1101/850529
Lopez SJ, Laufer BI, Beitnere U, Berg E, Silverman JL, Segal DJ, LaSalle JM. Imprinting effects of UBE3A loss on synaptic gene networks and Wnt signaling pathways. Human Molecular Genetics, 2019. doi: 10.1093/hmg/ddz221
Vogel Ciernia A*, Laufer BI*, Hwang H, Dunaway KW, Mordaunt CE, Coulson RL, Yasui DH, LaSalle JM. Epigenomic convergence of genetic and immune risk factors in autism brain. Cerebral Cortex, 2019. doi: 10.1093/cercor/bhz115
Laufer BI, Hwang H, Vogel Ciernia A, Mordaunt CE, LaSalle JM. Whole genome bisulfite sequencing of Down syndrome brain reveals regional DNA hypermethylation and novel disease insights. Epigenetics, 2019. doi: 10.1080/15592294.2019.1609867
This workflow is primarily based on the algorithms from the dmrseq and bsseq bioconductor packages. I would like to thank Keegan Korthauer, the creator of dmrseq, for helpful conceptual advice in establishing and optimizing this workflow. I would like to thank Matt Settles from the UC Davis Bioinformatics Core for advice on creating an R package and use of the tidyverse and also for help with the UC Davis example. I would like to thank Rochelle Coulson for a script that was developed into the PCA function. I would also like to thank Blythe Durbin-Johnson and Annie Vogel Ciernia for statistical consulting that enabled the global and chromosomal methylation statistics. I would like to thank Nikhil Joshi from the UC Davis Bioinformatics Core for troubleshooting of a resource issue. Finally, I would like to thank Ian Korf for invaluable discussions related to the bioinformatic approaches utilized in this repository.